Cancer is a disease of the genome and the epigenome that results from the sequential accumulation of genetic alterations. For the past few decades, enormous research has been performed to understand the genetic alterations involved in oncogenesis. This fundamental genetic understanding of cancer reveals its high heterogeneity and evolutionary dynamic (tumor evolution) as a function of time (Diaz and Bardelli, 2014; McGranahan and Swanton, 2017), particularly in response to treatment, thus not only highlighting the importance of genetics-based tumor analysis for precise management of cancer but also providing the potential for patient selection for targeted therapeutics. Early research was limited to tissue specimens obtained from invasive biopsy, surgical resections, organ transplantations, or upon autopsy. Recent advances in technology enable the study of cancer genetics using less invasive blood liquid biopsy without the need for tissue-based diagnostic tools (Crowley et al., 2013), and these advances have resulted in several liquid biopsy tests in clinic for cancer management (Clark et al., 2018; Janku et al., 2016; Kwapisz, 2017; Malapelle et al., 2017).
Noncell-associated DNA shed by either apoptotic or necrotic cells into circulation is referred to as cell-free circulating DNA (cfDNA). cfDNA shed by the tumor cells is referred to as circulating tumor DNA (ctDNA), and it can be a surrogate for the tumor genome. In the era of precision medicine, ctDNA is anticipated to play increasing role in diagnostics, targeted drug therapy/precision medicine, tumor response and recurrence monitoring, the development of anticancer drugs resistance, and so on.
Urine has been used with great clinical benefit as a source of reporter molecules for urine and nonurinary tract diseases (Blau et al., 1989; Bonn, 2004; Goldstein et al., 2004; Mangan, 2005). Urine-based tests are noninvasive and patient-friendly, and they can be used to monitor nonurinary tract site disorders such as diabetes and hypertension as well as other conditions, such as pregnancy (Suzuki et al., 1999; Goldstein et al., 2004; Mangan, 2005). Advances in molecular biomarker research and our previous findings that tumor-derived DNA in the circulation can be detected in urine (Botezatu et al., 2000; Iwaki et al., 2004; Quek et al., 2004; Su et al., 2004a, 2004b; Grossman et al., 2005; Lin et al., 2011; Song et al., 2012; Jain et al., 2015; Hann et al., 2017) have provided the opportunity to use urine as the biological fluid of choice to detect cancer-related molecular markers present in the circulation. Recent discoveries have established that DNA released from cells into circulation can be filtered through kidney into urine. This urine cfDNA can provide reliable and reproducible information on cancer-specific DNA alterations, a finding that is potentially useful for cancer diagnostics and monitoring. We previously reported that human urine contains DNA derived both from sloughed-off cell debris of the urinary tract and from the circulation (Su et al., 2004b). Then, we and others demonstrated that urine contains fragmented DNA that originates from organs outside the urinary tract and enters the circulation (Su et al., 2004a, 2004b, 2005, 2008a, 2008b), and urine is thus a viable substrate for detecting ctDNA markers (Su et al., 2008a; Song et al., 2012).
The use of circulating DNA for cancer detection has been studied extensively (Rogers et al., 1972; Chen et al., 1996, 1999; Anker et al., 1999, 2001; Castells et al., 1999; Anker, 2000; Gonzalez et al., 2000; Kirk, 2000; Kopreski et al., 2000; Jackson, 2001; Jahr et al., 2001; Anker and Stroun, 2002; Wong et al., 2003; Gautschi et al., 2004; Gormally et al., 2006; Pathak et al., 2006; Jiang and Lo, 2016; Wan et al., 2017; Cohen et al., 2018; Hench et al., 2018). Commonly, liquid biopsy refers to the use of blood as a minimally invasive source of body fluid for detecting ctDNA. However, urine collection, which is completely noninvasive, can serve as an alternate body fluid source. Given the direct access of the urinary tract with urine, considerable research and review has been focused on using urine for liquid biopsy of bladder cancer and other urological malignancies (Lin et al., 2017). In this study, we will focus on urine for genetic liquid biopsy of nonurological tract cancers.
Urine as a Body Fluid for Cancer Liquid Biopsy
Among available body fluids, blood is certainly the most direct body fluid for detection of ctDNA with most extensive studies on clinical applications in cancer liquid biopsy (Forshew et al., 2012; Crowley et al., 2013; Diaz and Bardelli, 2014; Franovic et al., 2017; Kwapisz, 2017; Malapelle et al., 2017; Pan et al., 2017; Wan et al., 2017; Hench et al., 2018). However, urine provides unique benefits as a truly noninvasive liquid biopsy; it can be collected, stored, processed, and shipped more easily than blood. Compared with blood, the collection of urine produces absolutely no discomfort to the patient and does not require trained medical staff. It is favorable for large collection volumes, which could overcome the diluted concentration of some ctDNA in urine. Urine also contains fewer contaminating proteins as compared with serum/plasma, making the process of DNA isolation easier. Furthermore, urine collection can be done at home and then shipped to certified laboratories for testing, thus eliminating the need for an in-person clinic visit and enabling more frequent monitoring. Urine can also be used more frequently in remote areas. Ultimately, the ease of use provided by urine-based liquid biopsy will enable the regular monitoring of high-risk populations, such as those infected with the Hepatitis B and C viruses or cirrhosis, for liver cancer. Overall, urine-based sample collection offers many advantages over blood, ultimately increasing patient compliance for cancer screening and permitting more frequent monitoring to enable patients to seek curative treatment and care.
Characterization of the Urine DNA
To explore the potential of using urine DNA as a source of cfDNA for cancer detection, we first characterized the urine DNA from different individuals. As shown in Figure 1A, nucleic acids isolated from the urine of five subjects were resolved into a common pattern displaying two distinct populations with respect to size: (1) a high molecular weight (HMW) form that remained near the well (H, 1 kb and larger) and (2) a low molecular weight (LMW) form (L, <400 bp) as detailed in (Su et al., 2004b). This ethidium bromide-stained nucleic acid was shown to be DNA, as it was sensitive to DNase digestion and resistant to RNase digestion (Figure 1B). The LMW urine DNA was mostly apoptotic in origin, as it co-migrated with 1–2 nucleosomal sized laddered DNA from serum (Figure 1C). Furthermore, through high salt wash and centrifugation, the HMW DNA was mostly cell-debris-associated, and the LMW urine DNA was cell-free soluble DNA in the supernatant (Sup), as indicated (Figure 1D). It is unlikely that cell-debris-associated HMW DNA can cross the kidney barrier, which means it likely arises from the cells shed into the urine from the urinary tract as well as from the small number of white blood cells that are detectable in urine. The nucleosomal laddering pattern of the LMW DNA suggests that it represents a portion of the circulating cfDNA from the bloodstream that has crossed the glomerular barrier, and the upper limit of its size is determined by the pore dimension in the glomerular barrier.
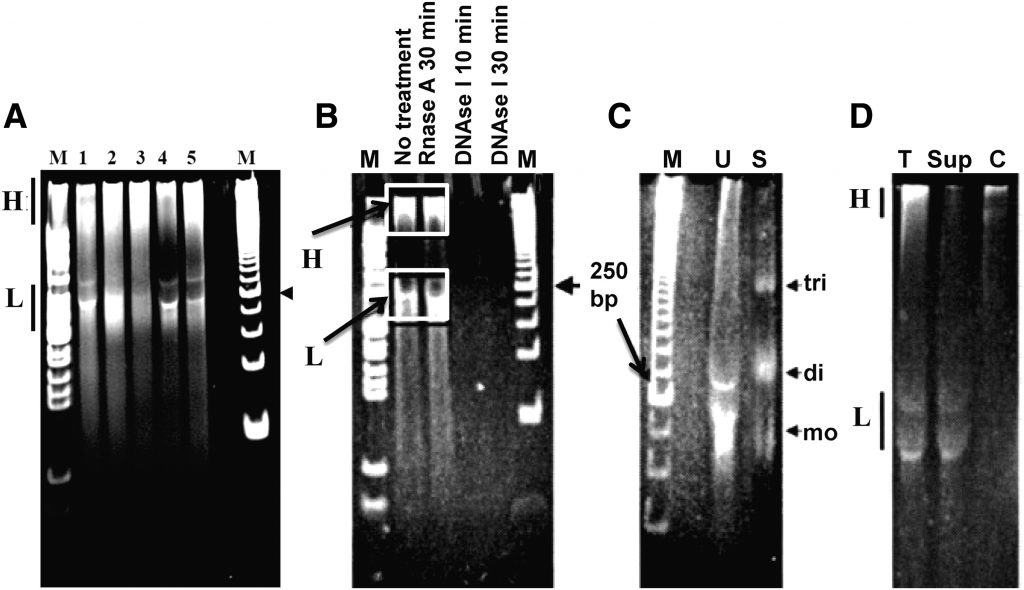
By using colorectal cancer (CRC) and hepatocellular carcinoma (HCC) as a study model, we further demonstrated the detection of ctDNA in the urine cfDNA LMW fraction. CRC-associated DNA modifications, such as KRAS mutations (Su et al., 2005, 2008a, 2008b), hypermethylation of the VIM gene (Song et al., 2012), and the HCC-associated TP53 codon 249 G > T hotspot mutation (Lin et al., 2011), were detected mostly in LMW urine DNA, but very limited, if any, in the HMW DNA fraction. Furthermore, a higher concentration of mutated KRAS DNA of the total KRAS (mutated and wild type) DNA detected in urine LMW DNA fraction as compared with that of total DNA (contains HMW and LMW DNA sizes) suggested that size selection for LMW DNA fragments could enrich for cfDNA, thus improving the sensitivity of detecting ctDNA markers. Collectively, this data demonstrates that the HMW urine DNA was mostly derived from the sloughed-off cell debris of the urinary tract, whereas the LMW urine DNA was mostly derived from the circulation that was enriched for tumor-derived DNA alterations if a tumor was present.
Approaches for the Enrichment of Urine cfDNA
As discussed earlier, ctDNA, if present in urine, is enriched in the LMW DNA fraction of the total urine DNA. Hence, separation of the LMW DNA or cfDNA fraction from the cellular or HMW DNA will enhance the sensitivity of the ctDNA detection. Two approaches have been used for this purpose: (1) centrifugation of whole urine to collect the supernatant and removal of cell pellet/debris and (2) size-based selection of total DNA for fractionation into HMW and LMW DNA. Commercial kits, such as the Extract-all Urine DNA kit (Zymo Research, CA), the Urine DNA isolation kit (Norgen Biotech, Thorold, Canada), the Genelute Urine cell-free DNA Urine mini kit (Sigma-Aldrich, MO), utilize the first approach based on urine centrifugation. We have developed the latter approach using solid-phase carboxylated magnetic beads (CMBs) to fractionate LMW and HMW DNA by varying the binding conditions, as detailed previously (Su et al., 2008a). Fractionation (or the removal of HMW DNA) leads to the enhancement of CRC-associated, mutated KRAS DNA detection (or ctDNA detection) from patients with CRC, as compared with using total urine DNA (Su et al., 2008a). No study has compared the two methods to indicate whether one is better than the other. However, the CMBs-based method offers the potential for automation for high-throughput specimen processing and eliminates the need for immediate urine processing compared with centrifugation.
Cancer-Related Genetic Alterations Detected in Urine cfDNA from Nonurological Tract Cancers
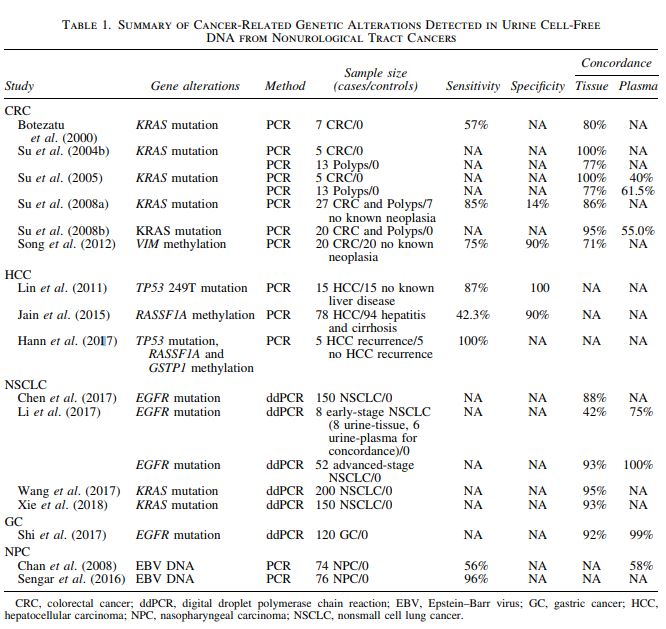
Genome-wide cancer-related genetic alterations, including mutations and methylation, have been reported in plasma-based liquid biopsy approaches across numerous cancer types. As compared with plasma cfDNA liquid biopsy, urine cfDNA has only been studied in a handful of cancer types, including CRC, HCC, nonsmall cell lung cancer (NSCLC), gastric cancer (GC), nasopharyngeal carcinoma (NPC), and breast cancer (BC). Table 1 summarizes the sensitivity, specificity, and concordance (with tissue and plasma), when available, of the detection of cancer-related genetic alterations in urine from these studies. The sample size of each study and the technologies used for detection are also listed in the table and detailed in the sections as follows. Note, either the highly sensitive polymerase chain reaction (PCR) or digital droplet PCR (ddPCR) were the methods used in these studies to detect the alterations of interest, implicating the limited amount of cfDNA in urine.
Colorectal cancer
Detection of CRC-associated genetic alterations, such as KRAS mutations (Botezatu et al., 2000; Serdyuk et al., 2001; Su et al., 2004a, 2004b, 2008a, 2008b), BRAF mutations (Janku et al., 2014), CAD–ALK gene arrangement (Siravegna et al., 2017), and methylation of the VIM gene (Song et al., 2012), have been reported in urine cfDNA. In fact, KRAS mutations in advanced CRC were the first reported biomarkers in urine from nonurinary tract cancers (Botezatu et al., 2000). Subsequently, Su et al. (2004b) demonstrated, in a small pilot study, 100% (5/5) concordance between mutant KRAS detection in urine and cancer tissue and 77% (10/13) concordance between urine and tissue from adenomatous polyps. Further studies established that the incidence of KRAS mutation in urine was similar to serum and plasma, but its incidence was significantly higher (95%) in urine than in either serum (35%) or plasma (40%) (p < 0.0005) when a larger amount of DNA, derived from 200 μL per body fluid per PCR reaction, was used, suggesting that inhibitory factors in serum/plasma may interfere with PCR assays (Su et al., 2008b). In addition to genetic mutations, methylation of the VIM gene could also be detected in the urine of patients with CRC (75% sensitivity, 90% specificity; 20 CRC, and 20 controls) using a 39 bp MethyLight qPCR assay to account for the small size of urine cfDNA and bisulfite-induced fragmentation of DNA (Song et al., 2012).
Recently, Siravegna et al. (2017) reported proof-of-concept data that urine cfDNA can be effective in monitoring tumor evolution in CRC. They detected the CAD-ALK gene fusion in the urine cfDNA of a metastatic CRC patient and studied the dynamics of the CAD-ALK rearrangement in plasma and urine, which were found to be concordant with and parallel to the patient’s clinical course. Detection of the CAD-ALK gene fusion in urine cfDNA anticipated radiological confirmation of disease progression.
Hepatocellular carcinoma
Using an LNA clamp-mediated 41 bp short amplicon qPCR assay, Lin et al. (2011) reported the successful detection of TP53codon 249 G > T mutation in the urine cfDNA of HCC patients with 86.7% (13/15) sensitivity and 100% specificity (0/15). Jain et al. (2015) reported the detection of methylated RASSF1A in urine in association with HCC and cirrhosis by using a 49 bp short amplicon methylation-specific PCR assay. Similar to CRC, a recent report by Hann et al. (2017) reported the detection of HCC-associated DNA markers, including the TP53 249T mutation and the aberrant methylation of the RASSF1Aand GSTP1 genes, for monitoring HCC recurrence. In this 10-patient study, the authors compared the detection of urine DNA markers with MRI scans for monitoring HCC recurrence. Five patients were confirmed by MRI for recurrence, and all five had detectable DNA biomarkers up to 9 months before MRI-confirmed recurrence, suggesting that detection of HCC-associated DNA markers in urine could provide a promising tool to complement detection of recurrent HCC by imaging.
Nonsmall cell lung cancer
A urine ctDNA assay was used to confirm the EGFR mutation and T790M resistance mutation in a case of NSCLC by Berz et al. (2016) to demonstrate the value of urine liquid biopsy as an alternative to tissue biopsy in a secondary resistance setting where tissue was not available for molecular testing and by Husain et al. (2017) for monitoring daily dynamics of early tumor response to anti-EGFR tyrosine kinase inhibitor therapy. A cost-of-care analysis study concluded that the urine testing strategy for detecting EGFR T790M resistance mutations prolonged progression-free survival (0.44 months) and overall survival (0.35 months) due to increased detection of T790M mutation and decreased biopsies and complication-related costs as compared with a tissue-testing strategy yielding an overall savings of $1243–$1680 per patient (Sands et al., 2017). Chen et al. (2017) also performed EGFR mutation testing using the digital PCR approach in both plasma and urine cfDNA. Urinary cfDNA had 88% concordance with EGFR mutation status with primary tissue and 98% concordance with plasma cfDNA. Similar results were obtained by Li et al. (2017). Mutant KRAS was detected in urine with 95% concordance with primary tissue biopsy (Wang et al., 2017) from an NSCLC patient cohort and longitudinal monitoring of urine specimens revealed association with disease progression and outcome (Wang et al., 2017; Xie et al., 2018) as well as the potential to assess therapeutic response (Tchekmedyian et al., 2017).
Gastric cancer
EGFR targeted therapy has been used to treat GC in patients with EGFR mutations. Shi et al. (2017) examined the potential of using urinary cfDNA as an alternative tumor material source for monitoring EGFR mutations in GC patients. They found a 92% concordance of EGFR mutations between urine and primary tissue at baseline and 99% concordance with plasma samples at different timepoints. This finding suggests that urine cfDNA is a reliable source for detecting EGFR mutations in primary GC and for predicting a patient’s prognosis, treatment response, and disease outcome. This is significant because obtaining repeated high-quality tissue biopsy from GC patients for monitoring the cancer status can be difficult.
Nasopharyngeal carcinoma
The gold standard for diagnosing NPC is the endoscopic examination of the nasopharynx and the histological examination of suspicious lesions, but it is costly and uncomfortable to the patients. Plasma Epstein–Barr virus (EBV) DNA analysis has been shown to be clinically useful in all aspects of NPC management, including detection, monitoring, and prognostication of NPC (Chan et al., 2003). Chan et al. (2008) first reported that circulating EBV DNA can be detected in the urine of NPC patients with positive correlation with its plasma concentration. Sengar et al. (2016) also compared EBV DNA levels in plasma and urine of patients with NPC and reported that urine EBV DNA has a high sensitivity (96%) at diagnosis, correlates well with plasma EBV DNA levels, reduces significantly with therapy, and that low EBV copy number was associated with improved survival.
Breast cancer
Urine cfDNA was recently utilized to explore the detection of PI3KCA mutations in BC (Liu and Liu, 2018). This study sought to evaluate whether liquid biopsy biomarkers, such as PI3KCA mutations in plasma or urine, could serve as a predictor of BC relapse in patients receiving therapy (Liu and Liu, 2018). In the 200 patients tested, they identified a positive correlation between plasma and urine DNA concentrations at baseline before the beginning of treatment. During plasma and urine serial sampling throughout the course of treatment, detection of PI3KCA mutations in urine and plasma DNA was found to be a potential predictor of relapse.
Conclusion
In spite of their many advantages in comparison with blood-based biopsies, urine-based biopsies have not been extensively developed or explored in nonurological cancers. Encouraging studies have recently demonstrated the great promise of the applications and clinical utility of urine-based liquid biopsies for cancer management. They can serve as a noninvasive approach to cancer screening, early detection, and frequent monitoring for recurrence, metastasis, and therapeutic efficacy. Furthermore, the molecular profiling of urine cfDNA can be prognostic and may reveal outcomes for patients that would otherwise be missed by standard clinical testing/surveillance. Similar to blood assays, preanalytical variables will need to be established for urine (i.e., collection, processing, downstream assessment) and more work is needed to verify and validate urine cfDNA biomarkers in larger patient cohorts. Overall, the noninvasive and patient-friendly nature of the urine-based biopsy warrants further development and offers a promising alternative to blood-based biopsies.
To access the original article in its entirety please see Urine-Based Liquid Biopsy for Nonurological Cancers.
Genetic Testing and Molecular Biomarkers, published by Mary Ann Liebert, Inc., is the leading peer-reviewed journal covering all aspects of human genetic testing including molecular biomarkers. The above article was first published in the April 2019 Liquid Biopsy special issue of Genetic Testing and Molecular Biomarkers. The views expressed here are those of the authors and are not necessarily those of Tissue Engineering, Mary Ann Liebert, Inc., publishers, or their affiliates. No endorsement of any entity or technology is implied.


