A review of approved drugs from 2020 and 2021 reveals that only 22% are designed against a truly novel target—a gene or protein that is thought to be capable of participating in a disease-modifying drug interaction. The implication, then, is that over three-quarters of recently approved drugs are not designed using new biological insights, despite two decades of mining the human genome.
The completion of the Human Genome Project in 2002 represented groundbreaking findings that significantly elevated our understanding of the human genetic instruction book.1 The human-focused project also paved the way for the analysis of genomes from other species, showing us that our genome and the other genomes in the animal kingdom are more similar than they are different, especially if the other genomes belong to closely related species, such as our fellow mammals.
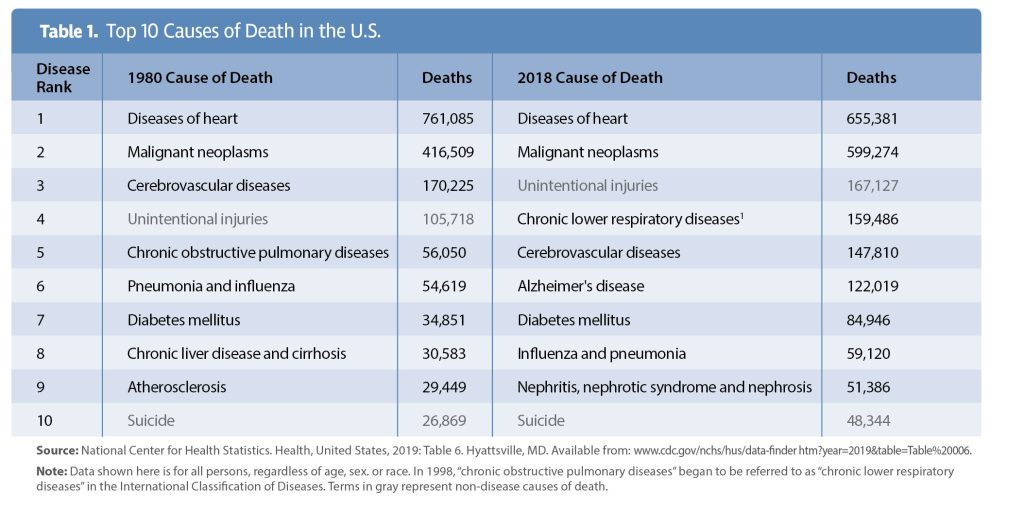
Flash forward 20 years. Despite the sequencing of over 30 million people, the top disease killers in the United States and worldwide remain largely unchanged.2 Heart disease, cancer, stroke, diabetes, respiratory disease, and renal disease kill over 1.5 million people each year in the United States alone (Table 1). Seven of the top killers in the United States—ischemic heart disease, stroke, respiratory disease, cancer, Alzheimer’s disease, diabetes, and kidney disease—are also top disease killers globally.3 Clearly, our approach of focusing almost exclusively on human data, along with a handful of imperfect model organisms, to drive innovation and support the development of new treatments for these diseases is flawed.
Innovative technologies and approaches are needed to improve our understanding of susceptibility and to inform our efforts to discover novel targets. One approach is to look beyond traditional laboratory rats and mice, which are used to crudely approximate disease in humans, and instead focus on disease avoidance strategies that have evolved in other animals, some of which could hold the secrets to unlocking protective mechanisms that already exist humans but are inactive.
Comparing genomics across species
Evolution in the genome tends to operate on the “if it’s not broken, don’t fix it” principle. The parts of the genome essential for survival and reproduction tend to remain the same over millions of years. When we compare genomes of different species, we see these sequences are very similar (or conserved). Identifying a change in a conserved part of the genome that is specific to one species or a set of species with a given characteristic can reveal the genomic basis of that characteristic.
For example, naturally low biliary phospholipid levels in guinea pigs and horses have been linked to the gene Abcb4 by analyzing common patterns of genetic variation. Similar mutations in the human version of the gene, ABCB4, also result in low biliary phospholipid levels but also severe liver disease.4
Comparative genomics also shows that for some diseases, these types of modifications inevitably lie in the part of the genome that does not encode for genes, as genes are generally highly or even entirely similar between species. Therefore, integrating epigenomic data (or understanding how genes are regulated) will improve our understanding of disease and prove essential for genome-based therapeutic discovery in the future.
Natural disease resistance is a phenomenon where species have evolved mechanisms to either prevent a disease entirely or, perhaps more interestingly, respond to the disease after it has started to develop. For example, after aspects of a disease develop in organisms of such species, those organisms may be able to reverse those aspects and return to a state of health.
This can be seen in humans as well, in families that inherit specific genetic mutations that protect them from disease (such as in the case of mutations in the gene PCSK9 resulting in low cholesterol).5 There are companies now mining specific human populations for these aberrations. Variant Bio is one such company examining specific populations of humans with extreme traits (including obesity or resistance to certain infectious diseases) to search for new drug targets. Unfortunately, as a whole, humans are just too similar, and there are not enough naturally occurring human populations resistant to the many complex diseases that ail us.
But there are many examples of this when you examine the broader animal kingdom.
Studying disease resistance in other species
The 13-lined ground squirrels and tenrecs naturally resist strokes and heart attacks when they drastically reduce the oxygen supply in their brain and heart during hibernation-like states called torpor. Such an experience would cause inflammation and fibrosis in humans, but these consequences are entirely avoided in these species.
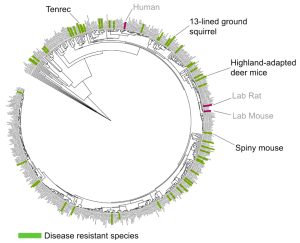
Evolved disease avoidance in mammals is not unique to hibernators. The spiny mouse (Acomys cahirinus) seemingly regenerates skin and multiple organs (including the kidney and spinal cord) without fibrosis or scarring after injury. Out of the 300 species reviewed by Fauna Bio thus far, 51 species have been found to have evolved resistance toward one or more human disease states.
Following genetic clues
Multiple studies now support the assertion that drugs with genetic support are more likely to succeed in clinical trials6,7 and be approved as therapeutics. However, we can improve our ability to find functional genetic targets by understanding which genes have been highly conserved across evolution, as these genes are more likely to contribute to disease, particularly for the genetic risk of complex diseases (including diseases such as coronary artery disease and type 2 diabetes).8
It makes sense that genes that have not changed in hundreds of millions of years often do something meaningful. Improved sequencing technologies have enhanced our understanding of genetic conservation across hundreds of species. Combining this with functional data from species that are using those genes in unique and protective ways provides us the ability to rapidly home in on those genes that are highly functional and can, hopefully, help us prevent or reverse disease in humans.
Scientists, physicians, and even the National Human Genome Research Institute acknowledge that there is much more to do, and that the human genome alone doesn’t hold all the answers we need.9 By continuing to innovate, investigate, and build on the available data we have today, we can open up many new potential classes of targets and medicines that will impact some of the biggest known killers of humans today.
Ashley Zehnder, DVM, PhD, is chief executive officer of Fauna Bio.
References
- National Human Genome Research Institute. Human Genome Project Results. Last updated: November 12, 2018. Accessed December 5, 2022.
- Crespi S. Looking back at 20 years of human genome sequencing [podcast]. Science. Posted February 4, 2021. Accessed December 5, 2022. DOI: 10.1126/science.abg8636.
- World Health Organization. The top 10 causes of death. Posted December 9, 2020. Accessed December 5, 2022.
- Hiller M, Schaar BT, Indjeian VB, et al. A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2012; 2(4): 817–823. DOI: 10.1016/j.celrep.2012.08.032.
- Benton ML, Abraham A, LaBella AL, et al. The influence of evolutionary history on human health and disease. Nat. Rev. Genet. 2021; 22(5): 269–283. DOI: 10.1038/s41576-020-00305-9.
- Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015; 47(8): 856–860. DOI: 10.1038/ng.3314.
- King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019; 15(12): e1008489. DOI: 10.1371/journal.pgen.1008489.
- Finucane HK, Bulik-Sullivan B, Gusev A, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015; 47(11): 1228–1235. DOI: 10.1038/ng.3404.
- National Human Genome Research Institute. Comparative Genomics Fact Sheet. Last updated: August 15, 2020. Accessed: December 5, 2022.

