December 1, 2016 (Vol. 36, No. 21)
Seemingly Minor Communications Glitches or Security Breaches Can Derail Ordinarily Smooth Operations
Cell signaling systems—the molecular mechanisms that cells use to maintain homeostasis or respond to crisis—work in accordance with intricate and delicately calibrated rules. Unfortunately, the rules can be altered, such that cell signaling continues to “work,” albeit to perverse ends such as autoimmunity or cancer. But if the rules can be changed for ill, leading cells astray, perhaps they can also be changed for the better, guiding cells back to their proper, healthful routines.
Therapeutic revisions to cell signaling will be possible only if we become conversant in the language of cell signaling, which is not so simple as a code, such as semaphore, Morse code, or radio operator’s lingo. It is even more complicated than the signaling system that manages the telephone network, setting up and tearing down calls and passing on metadata.
In all these human-made signaling systems, the signal is distinct from the signal-dependent communication, a voice session, for example, or more substantively, the signal-dependent transfer of cargo or passengers.
Cell signaling is more like Internet signaling, where routing and priority information is embedded with the substance of what is being routed and prioritized. Granted, molecules are not packets. They lack “headers” and “trailers” that contain control information. They can, however, be phosphorylated, acetylated, or methylated. Such in-band signaling can realize a cellular version of the “Internet of things,” except in this case the things are molecules. If such signaling becomes corrupted, a security patch may be available.
Deciphering Cancer Signatures
Characterizing cell-signaling networks at the intersection of multiple pathways can reveal myriad complex interactions, especially in cells transformed by cancer. According to a report credited to scientists based at Cell Signaling Technology (CST), a proteomic approach can identify new druggable disease markers by comparing normal versus malignantly transformed tissues.
This report, which was presented to the 2016 American Association for Cancer Research meeting, describes a study of lung cancer, the most common type of cancer in the United States. In particular, the report focuses on small cell lung carcinoma (SCLC), which has a dismal survival rate of 6%.
“Our goal was to develop a highly quantitative tandem mass spectrometry (MS/MS) approach coupled with tandem mass tag (TMT) labeling and specific antibodies to identify deregulated pathways between tumor and normal specimens,” says one of the report’s co-authors, Klarisa Rikova, a researcher who recently left CST to join Bluefin Biomedicine.
According to Rikova, the CST team utilized motif antibodies to post-translational modifications (PTMs) for immuno-isolation followed by mass spectrometry (MS): “Often, PTMs are deregulated in cancers. These pathways include phosphorylation, acetylation, and methylation that act alone and in combination in order to regulate protein function, cellular behavior, and epigenetics.”
The scientists digested tissue with trypsin, labeled with TMT reagents, serially fractionated by immunoprecipitation with PTM-specific antibodies, and performed a six-plex TMT MS analysis. Rikova notes, “We identified more than 15,300 differentially phosphorylated, acetylated, and methylated sites on over 4,600 proteins from 17 SCLC patients’ tissues and 5 paranormals.”
Rikova envisions that this approach can be utilized for many other types of applications including preclinical assessments and evaluations of patient responses to new drugs. “The real challenge with generating this amount of information is how best to utilize and understand the data. The CST team hoped to generate results that could be examined by others and yield interesting comparisons.” The CST team also had in mind the development of proteomic approaches that could be used by the company’s clients.
“It is amazing how much information one can generate,” concludes Rikova. “The challenge is always assigning function to findings.”
Cannibalizing Cancer Cells
Entosis is a fascinating act of cellular cannibalism in which one live (and apparently viable) epithelial cell is engulfed, killed, and digested by another. Entosis comes from the Greek word “entos,” which means inside or within. Over the past 100 years, pathologists have conducted studies of this process, and they have been particularly interested in how it unfolds within human tumor specimens. Entosis utilizes a complex set of signaling mechanisms that may be subverted by cancer.
The process of entosis is a double-edged sword for cancer, comments Joanne Durgan, Ph.D., a postdoctoral researcher at the Babraham Institute: “On one hand, the murder and digestion of one cancer cell by another has the capacity to confer tumor suppressive activity, effectively limiting tumor outgrowth.
“On the other hand, as the remaining cell gains valuable nutrients from cannibalizing its neighbor, this transaction simultaneously confers a survival advantage to the host that may be of particular benefit in the nutrient-deprived conditions of a tumor.
“In simplified terms, entosis may be viewed as growth suppressive from the perspective of the inner cell, but growth promoting for the host. Further work will be required to define the overall impact and significance of this process.”
Part of Dr. Durgan’s work focuses on the role of entosis in cancer biology. “We are exploring the molecular mechanisms that underlie this process and seeking to understand its impact on tumor progression,” she explains. “We also study entosis from a broader cell biological perspective, using it as a model with which to explore cell engulfment and lysosomal degradation.”
Dr. Durgan notes that entosis represents a suitable model for studying the emerging link between noncanonical autophagy and cellular engulfment events. “The large entotic vacuole is uniquely amenable to microscopic analyses and imaging-based screens,” she emphasizes. “In our ongoing work, we are using entosis to learn more about the fundamental mechanisms of noncanonical autophagy signaling, vacuole processing, and lysosomal recruitment that are at play during a broad range of macroendocytic engulfment events.”
Dr. Durgan is also considering how signaling pathways might influence cell contractility: “We are currently studying this aspect of entosis in collaboration with biophysicists. Signaling pathways that influence contractility determine the inner/outer cell identities, conferring a means of cell competition through entosis. It is known that oncogenic Ras controls entosis by influencing the biophysical properties of the cell, and it will be of interest to determine which other oncogenes and associated signaling pathways can regulate cell cannibalism in this way.”
Outsmarting Cancer
Tumor cells often overexpress key proteins that help them avoid recognition by host cells seeking to eliminate them. One such example is CD47, a ubiquitously expressed transmembrane protein that partners with membrane integrins and binds to thrombospondin-1 and signal regulatory protein alpha (SIRPα).
“CD47 has a fascinating biology in that it binds to SIRPα on macrophages and delivers a ‘do not eat’ signal,” notes Bob Uger, Ph.D., CSO, Trillium Therapeutics. “In the past five years, data in the cancer field demonstrate that increased expression of CD47 on tumor cells often correlates with poor clinical outcome.”
According to Dr. Uger, the company’s approach is to block this signal using a decoy receptor. “We’ve developed SIRPαFc, which is a fusion protein consisting of part of SIRPα, the CD47 receptor, linked to the Fc domain of an immunoglobulin that activates macrophages via Fcγ receptors. Thus, when administered, SIRPαFc binds to CD47 on cancer cells and prevents them from delivering the suppressive ‘do not eat’ signal.
“Furthermore, increased phagocytosis of cancer cells can translate into a better T-cell response. Thus our SIRPαFc may activate both innate and adaptive immunity.”
Trillium Therapeutics has begun two multicenter Phase I trials in patients with advanced hematological malignancies and solid tumors. While SIRPαFc is given via intravenous infusion in the hematological malignancy trial, the company is employing intratumoral injections in solid cancers.
“We distinguish our approach from others,” asserts Dr. Uger. “And we do it in a particularly important way: SIRPαFc does not bind red blood cells, thus reducing the risk of anemia and also potentially avoiding the large sink effect.”
For the future, Dr. Uger sees the possibility of utilizing SIRPαFc in combination with other chemotherapeutic agents. “We are considering all potential options,” he explains. “We are very excited about the future prospects for this therapeutic approach.”
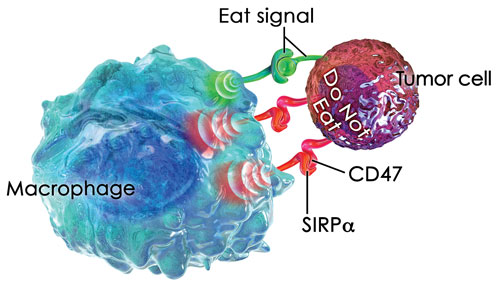
Trillium Therapeutics’ lead drug candidate, TTI-621 (SIRPaFc), targets CD47, a “do not eat” signal that tumor cells exploit to escape destruction by the innate immune system.
Hunter of Histidine Phosphorylation
Many proteins have one or more phosphorylation sites involved in signaling. The majority of intracellular proteins are phosphorylated at any given time. Although 9 of the 20 amino acids can be phosphorylated, focus has centered primarily on serine, threonine, and tyrosine phosphorylation. However, histidine phosphorylation is emerging as an important player in this complex network of phosphosignaling.
“New mass spectrometry–based phosphoproteomic studies have identified thousands of phosphorylation sites in human cells, tissues, and tumors,” says Tony Hunter, Ph.D., professor, Salk Institute. Dr. Hunter, known affectionately as the “King of Kinases,” made the seminal discovery more than 30 years ago that tyrosine kinases could add or subtract phosphates and thereby activate or deactivate signaling proteins. Further, he demonstrated when this aspect of the signaling system malfunctions, as it does in cancers, the growth of cells is switched to an ‘always on’ mode.
Among other things, Dr. Hunger is currently targeting phosphorylation of histidines, a lesser known phenomenon. “Early indications suggest that phospho-histidine (pHis) could be just as prevalent and important as the better known tyrosine phosphorylation,” he notes.
“Although pHis has been extensively studied in bacteria, its role in mammalian signaling remains largely unexplored because of the lack of specific antibody tools as well as the lability of its phosphoramidate bond. The latter makes it unsuitable for conventional mass spectrometry and other types of analyses.”
Dr. Hunter and colleagues decided to take on the difficult task of developing monoclonal antibodies to the 1- and 3- isoforms of pHis. “It took us about four years, but we developed sequence independent monoclonal antibodies to pHis,” he reports. “They do not bind to phosphotyrosine and are able to distinguish the two forms of pHis (depending on which of its two imidazole nitrogens are phosphorylated). We utilized these monoclonal antibodies to identify pHis substrates and functional sites using a variety of immunological, proteomic, and biological assays.”
According to Dr. Hunter, his group initially identified 786 enriched proteins. “This serves as a starting point for analysis,” he advises. “The complete list will likely be substantially longer. However, the analyses revealed a wealth of findings. It appears that pHis signals may be important for regulating multiple cell-cycle events.
“Also, this includes multiple interaction nodes involved in processes such as cytokinesis, chromosome condensation, and chromatin and spindle organization. Further, gene ontology analysis has implicated processes such as nucleic acid-related processes, RNA processing, and RNA splicing.”
Dr. Hunter describes a recent study taking advantage of the new monoclonal antibodies as well as other technologies: “In collaboration with Edward Skolnik, M.D., Ph.D., director of the division of nephrology at New York University Medical School, we showed that phosphorylation of His358 in the cytoplasmic tail of the potassium channel protein KCa3.1 activated ion flux by antagonizing copper-mediated inhibition of the channel. The metal-binding properties of histidine and its ability to become phosphorylated distinguish it from phosphorylation of serine/threonine and tyrosine.”
These findings also have therapeutic implications because limiting intracellular copper can enhance activation of T cells, thus opening the door to enhance Tcell–mediated killing of certain types of tumors via the enhanced activation of KCa3.1.
“There are many challenges remaining in the study of histidine phosphorylation,” concludes Dr. Hunter.
“We and others continue to address many technical hurdles such as the need to create better methods to study pHis using mass spectrometry. There are also exciting possibilities in the arena of genetic analyses, which may, for example, help us determine how pHis affects cell cycle progression. Such insights would be especially welcome in studies of cancer development.”
Cytokine Profiles Shown to Correlate with Autism Severity
The causes of autism spectrum disorders (ASDs) are poorly understood despite intense research on the disease around the world. One research group is taking an out-of-the-box approach to probe a decades-old connection between the disorder and the immune system.
A team at University of California, Davis, led by Judy Van de Water, Ph.D., a professor in the department of internal medicine, is investigating whether the intriguing link between ASD and immune system anomalies is causal or coincidental. In particular, Dr. Van de Water’s laboratory focuses on the immune system’s influence on neurodevelopment during pregnancy, at birth, and during early life. She is approaching the problem by studying the relationship between the cytokine profiles in children with ASD and the severity of their disorder.
Creating a snapshot of the immune response requires analysis of dozens of cytokines. However, the UC Davis researchers work with precious samples—either neonatal blood spots or blood drawn from very young children where the developing immune system is immature and cytokine production is low. Cytokine analysis under these conditions can prove quite challenging.
Traditional ELISAs can work with only one analyte at a time, which would necessitate dozens of experiments for each sample. This is not ideal when dealing with precious samples, as the amount of starting material would simply not be enough. The researchers therefore used a Bio-Plex system (Bio-Rad Laboratories) with Luminex xMap technology, which employs a mixture of color-coded beads that can detect and quantify multiple cytokines simultaneously. To reliably quantify cytokine expression with limited sample, the team also used highly sensitive Bio-Plex Precision Pro Human Cytokine kits with a low limit of detection.
Aided by the multiplex system and cytokine kits, Dr. Van de Water’s team identified several cytokines associated with severity of ASD. Her research could help develop immune-based diagnostics for ASD, eventually resulting in earlier diagnosis and more effective treatment.




