January 1, 2012 (Vol. 32, No. 1)
Bioanalytical preparation is at the core of drug discovery and the clinical decision-making process. This area has seen many technological breakthroughs in instrumentation as well as multiple advances in analytical techniques.
Novel technologies aim to isolate the target component from complex biochemical samples, shorten the time from sample collection to diagnosis, and deliver as-yet undiscovered targets. Experts in sample-preparation technologies for detection, identification, and analysis of biomolecules explored the latest R&D developments as well as ready-to-market technologies at Knowledge Foundation’s “Integrating Sample Preparation” conference held last month.
When cancer cells undergo necrosis, their nuclear DNA, organelles, and other cellular debris end up in the bloodstream. The ability to rapidly detect circulating cell-free cancer DNA would be a major advance in early cancer detection and screening.
As opposed to short pieces of DNA generated during the course of normal apoptosis, DNA particulates from necrotic cancer cells generally have high molecular weight (hmw). Therefore, appearance of hmw-DNA can serve as a biomarker of the cancer development, especially at early stages, when over 90% of cancers could still be successfully treated.
Low levels of hmw-DNA present considerable challenges for assay development. Conventional sample-preparation technologies result in significant losses of hmw-DNA, limiting the use of this important biomarker.
“A seamless sample-to-answer diagnostic system is critical when analyzing low-quantity biomarkers in a high-noise background,” said Michael J. Heller, Ph.D., professor of bioengineering and nanoengineering, University of California, San Diego.
“Dielectrophoresis (DEP) is a technology capable of separating cells and bacteria as well as nanoscale objects, so it works in the correct molecular size range. However, DEP applications were previously limited by the fact that separations could only be carried out under low conductance conditions.
“Blood and other biological fluids, which have relatively high conductance, had to be considerably diluted for DEP separation. Dilution results in loss of already rare entities such as circulating cancer DNA biomarkers.”
DEP induces the motion of particles in the alternating current using electrodes of different sizes or electrodes in asymmetric arrangements. Migration of molecules depends on the dielectric differences between the molecules and the media. As a result, some particles aggregate in the DEP low field regions and some migrate into DEP high field regions.
Dr. Heller’s lab identified a set of conditions within narrow brackets of AC electric voltage and frequency that enabled DEP separation of biological nanoparticles in high ionic strength (high conductance) samples, such as blood, buffy coat blood, plasma, or cerebrospinal fluid.
“Our proof-of-concept experiments were done using platinum microelectrode arrays overcoated with a hydrogel layer. While most DEP electrode devices simply disentegrate under high conductance conditions, our devices held just enough to show DEP separation,” continued Dr. Heller.
Biological Dynamics, a start-up from Dr. Heller’s lab, has subsequently improved the technology, creating electrodes capable of withstanding high ionic dielectrophoresis conditions. The company is perfecting the technology with additional capabilities such as PCR or sequencing directly from the array.
Low-Tech Sample Prep
“Molecular diagnostic tools have grown to be very sophisticated,” said Season Wong, Ph.D., senior scientist, Lynntech. “At the same time, these tools tend to rely on the availability of robotic systems, or at least centrifuges. However, the need for rapid molecular tests has risen dramatically in low resource settings, where even electricity may not be available.”
NASA has collaborated with Lynntech to develop a sample-preparation system capable of working in zero gravity. The increased time lag between sample collection and its analysis on Earth also increases the chance of sample degradation. Immediate analysis at the space station may become critical to preserve and expand our knowledge of the universe.
“Moving liquids in zero gravity is difficult,” contended Dr. Wong. “In Lynntech’s system, liquids remain static. Magnetic forces move only the target nucleic acids.”
The sample-prep system consists of several chambers, filled with lysis, wash, and elution buffers in a customized order. Chambers are connected by plug valves. A sample is lysed and nucleic acids are captured on magnetic beads.
When a magnet rod is inserted into the plug valve, the beads are attracted to the valve and accumulate in a dimple on the side of the valve. When the valve rotates 180°, the beads rotate with it. Removal of the magnetic rod deposits the beads into the next chamber. The sample passes through chambers until it is eluted from the beads and used in the analysis.
The system can be actuated manually or automatically, performing a typical nucleic acid purification scheme within seven minutes.
“Since the system is fully closed, cross-contamination is minimized,” added Dr. Wong. “This feature is particularly important when testing for highly infectious agents, particularly in low-tech environments and remote areas.” Lynntech is also developing the sample-preparation technology for molecular diagnostics in nontraditional health settings.
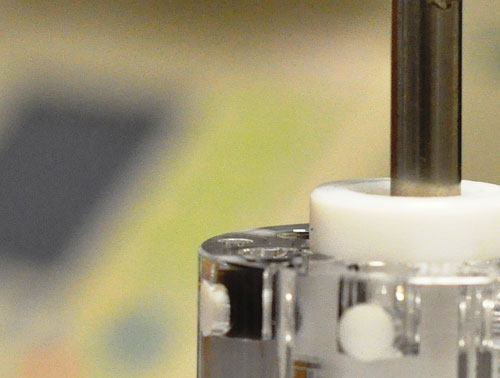
Working with NASA scientists, Lynntech has developed a sample-prep system capable of working in zero gravity. In this system, liquids remain static, and magnetic forces move only the target nucleic acids.
“Dirty” Samples
Molecular analysis typically involves a priori knowledge of the target bioparticles or biomolecules. However, many applications would benefit from a more generic approach capable of separating all bioparticles in a given sample, including unidentified targets.
“Complex real-life samples often present a large background of cellular debris and host genome,” said engineer Maxim Shusteff, leader of the microfluidic sample-preparation effort at Lawrence Livermore National Labs.
“Our challenge is to design a modular system capable of separating bioparticles by physical properties such as size, density, or mobility under electric fields, as opposed to affinity. Specifically, the system should be able to deliver viral pathogens that may be present in a low titer.”
The prototype instrument contains four plug-and-play modules. The modules serve as filters, retaining and passing particles of a certain size. Each module is built around a microfluidic chip, in which two fluids flow side-by-side down a microscale channel.
Because of viscosity at microfluidic scale, there is no mixing of the sample and recovery fluids, while fractionated particles are able to transfer between the fluid streams. The modules running acoustic focusing, dielectrophoresis, isotachophoresis, and thermal lysis may be linked in any configuration required by a particular application.
“Acoustic focusing is the most robust module currently being validated with real forensic samples,” continued Shusteff. The team uses this technology to rid the sample of cellular debris and bacteria (>2 µm in diameter) while retaining viral particles (typically 20–200 nm in diameter).
An ultrasound standing wave is generated by vibrations of a piezoelectric transducer, creating areas of high and low pressure. In this module, the area of minimum pressure creates a plane within the recovery channel. Larger particles concentrate single-file along this plane and are carried out by the recovery fluid.
To increase the size of the resonant cavity, the team added a bypass channel. The ultrasound wave transmits through a partition between the bypass channel and the adjacent microchannels. Viruses are too small to be influenced by the acoustic waves, and continue on to the next module.
The module is capable of recovering more than 80% of viral particles and removes more than 90% of cells from the test samples, Shusteff claimed. This compares favorably with time-consuming manual filtration, currently the standard method.
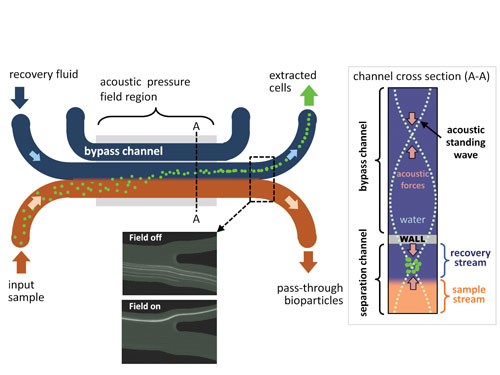
Lawrence Livermore National Lab’s “through-the-wall” acoustic focusing device allows for the precise location of a focused stream of cells in a recovery fluid. Smaller particles (
Standardizing Evaluation
Shortening the time of molecular-based detection assays requires rapid DNA extraction from a variety of real-life samples such as soil, food, and tissues. Performance of DNA extraction methods vary widely based on the nature of the sample. Quantity and quality of the resulting DNA are two parameters used to evaluate the performance of the extraction method.
“We have developed a standardized process for assessment of any given DNA purification method,” said Nathan Olson, research scientist, National Institute of Standards and Technology. “These standard procedures guide the decision as to which DNA extraction method is the best for this particular sample type.”
The team evaluated the efficiency of six different DNA extraction methods using five organisms including Gram positive and negative bacteria, spores, and yeast cells. The resulting 113 DNA preps were methodically scored based on DNA concentration, purity, and fragmentation.
Extraction efficiency was calculated using two methods. First, by the ratio of DNA yield determined by fluorometric measurements to estimated starting DNA; and second, by the ratio of a marker gene copy number determined by quantitative PCR (qPCR) to the number of cells extracted.
The observed extraction efficiency and DNA integrity varied by both organism and extraction method. “Using such matrix analysis for evaluation of assay performance is especially critical when testing new starting complex material,” continued Olson.
“For example, mixed microbial communities are analyzed as a pool. Qualification of the extraction method would enable a researcher to determine whether the method itself skews the gene-expression results in favor of the organism particularly suitable for this method.”
The team is working on establishing consistent standards for mixed bacterial communities that can be used for characterization of DNA-extraction methods.
Macro to Micro Volumes
Detection of very low numbers of organisms in large volumes of liquid necessitates some form of enrichment prior to further processing. InnovaPrep technology concentrates trace particles from liters of starting material to as little as 50 µL.
The system separates particles from the fluid using porous hollow fibers. The fiber “straws” are made out of membranes with precision pores. Depending on application, pore sizes from 5 nanometers to 20 microns can be used.
With one of the ends of the fiber straw blocked by a valve, the sample is forced into the center bores of the fibers. Liquids are vacuum-filtrated through the fiber pores, and the particles sediment inside the fiber straws.
“PCR and sequencing reactions are very sensitive to the presence of metal ions and inhibitor proteins,” said David Alburty, CEO.
“Concentration combined with buffer exchange during the process minimizes this interference by reducing unwanted material. However, if it is free DNA you are after, the permeate, now free of particular matter, can be concentrated again on fibers with smaller pores.”
An elution method used by all InnovaPrep Concentrators enables maximum recovery of the retained material in the desired volume of the recovery buffer, compatible with downstream applications. The recovery buffer is carbonated, generating wet foam that is expanded up to ten times the starting volume.
The viscous foam is released through the bore of the fibers, scraping the particles off the insides. As soon as it comes out from the now open ends of the fibers, the foam collapses into flat liquid, which can be pumped straight into PCR instrumentation.
“Our HCI-40 Integratable concentrator is designed to be a connecting link between various existing sampling systems and DNA analysis systems,” said Alburty.
InnovaPrep collaborated with BioWatch, a national surveillance program designed to provide early detection of biothreats.
A BioWatch Gen3 system, integrated with the Innovaprep Concentrator, continuously sampled outdoor air over a period of several months. This combination enabled near real-time reporting of airborne pathogens and provided two orders of magnitude performance improvement, according to Alburty.

InnovaPrep’s Integratable Sample Con-centration Unit HCI-40 was built for ease of integration into sample collection/detection systems and possesses the ability to separate nucleic acids and proteins from viable cells and concentrate the target organisms or proteins for improved detection, according to the company.



