March 15, 2012 (Vol. 32, No. 6)
Steady progress in a variety of technologies, such as gene-expression profiling, proteomics, and next-generation sequencing, is helping to characterize tissues and cells and to catalog the differences between healthy and diseased tissues. The ability to go beyond these techniques and to analyze and monitor the status, activity, and features of individual cells is yielding information on why cells of the same type may respond differently to external stimuli or therapeutic compounds, how tumor heterogeneity may affect drug response and secondary drug resistance, and how surrounding microenvironments in vivo or in bioreactors can be manipulated to alter the growth and function of cells and microbes, for example.
A variety of tools and techniques are in use to enable single-cell analysis, including microfluidics and other methods to isolate single cells in controlled microenvironments, microfabrication techniques and nanoprobes to create cellular environments in vitro, and miniaturization technology to enable assays and molecular detection at the single-cell level.
At the upcoming Select Biosciences “Single Cell Analysis Europe” conference, Birgitta Knudsen, Ph.D., associate professor, department of molecular biology and genetics at the Interdisciplinary Nanoscience Center (iNANO), Aarhus University, Denmark, will describe a microfluidics-based molecular detection approach for measuring enzyme activity in single cells that can achieve multiplexed detection of individual enzymatic events.
The technology underlying this approach is isothermal rolling circle amplification (RCA), an established method for direct detection and quantification of nucleic acid sequences.
Dr. Knudsen’s group has developed a rolling-circle-enhanced enzyme activity detection (REEAD) assay. This assay is based on RCA technology that allows for detection and visualization of enzymatic DNA cleavage-ligation events using fluorescent probes and provides single-molecule sensitivity. REEAD can measure the activity of an enzyme and not merely its presence in a cell.
The initial application of this approach is to detect topoisomerase I (topI) activity in human cancer cells. TopI, a DNA-cleaving enzyme, is an important target for anticancer drugs used to treat ovarian, colon, and small-cell lung cancer, for example.
Approximately half of the patients do not respond to the treatment, however, and in more than 50% of patients secondary drug resistance develops after an initial response to chemotherapy. The reason for this is unclear, but it is known that average topI levels across a tumor cell population correlate to drug response. High topI activity is associated with a good drug response.
One of Dr. Knudsen’s research goals is to explore how individual cancer cells vary and how that variation affects cell response to chemotherapy. She then envisions being able to apply that knowledge to the analysis of tumor tissue from biopsy samples and to use markers such as topI activity to help clinicians predict drug response and guide drug selection.
Frederik Fritzsch, a researcher in the laboratory of chemical biotechnology at the department of biochemical and chemical engineering of the TU Dortmund University (Germany), will discuss two key terms: “controlled” and “contactless.” Why are these so important for single-cell analysis?
“The principal reasons for phenotypic diversity and heterogeneity in bioprocesses with isogenic populations are the varying microenvironments surrounding single cells,” says Fritzsch. The ability to control this microenvironment allows scientists to introduce systematic perturbations and perform time-resolved analyses to assess the effects of these perturbations on single cells.
The results can be used to decipher cellular mechanisms, biochemical pathways, and regulatory cascades, and may lead to the identification of biochemical targets for drug discovery. They can also facilitate metabolic engineering of hyper-producing microorganisms for industrial biotechnology applications.
The ability to control the microenvironment of a cell implies control over both physical (including temperature and mechanical impact such as pressure or shear stress) and chemical (by modifying the media and nutrient supply) parameters.
“Contactless” refers to the trapping of a single cell such that it does not come in contact with surfaces or other cells. Noncontact isolation of a cell is important because cells intrinsically interact with their surroundings, resulting in potential changes in their morphology, function, signaling pathways, and surface features.
Established lab-on-a-chip-based methods for performing contactless single-cell analysis typically use negative dielectrophoresis (nDEP) to trap cells. These approaches have limitations, including Joule heating; in addition, they are also not suitable for capturing cells
Simulating Cellular Microenvironments
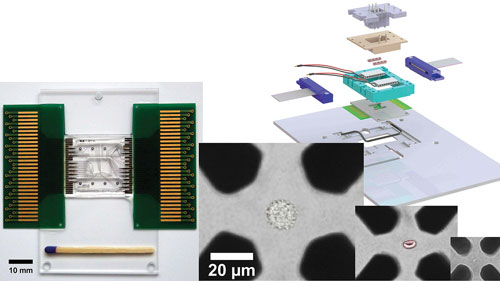
In his talk, Fritzsch will describe his work in the laboratory of Andreas Schmid, Ph.D., to develop an nDEP chip designed for use in the microfluidic chip-based cell analysis system called the Envirostat 2.0.
“Computational modeling and cell experiments were used to optimize a microelectrode and microchannel design” that allows for precise control of the microenvironment surrounding an individual cell, including small cells and bacteria, and enables, for example, the isolation of a single bacterium from a fermentation broth and removal of media samples for analysis.
Once trapped in a media stream, a cell or organism can be analyzed using a variety of methods, including direct growth analysis, use of fluorescent reporter systems, and collection of secreted molecules for analysis using optical or mass spectrometry techniques.
These methods can be used to study single cells derived from a variety of human cell lines and microbes, and make it possible to separate and analyze mother and daughter cells in parallel. For process design and optimization studies, Fritzsch describes the potential to isolate a single cell or bacterium and simulate the environment in a bioreactor system.
It is then possible to modify one or more of the physical or chemical parameters and study the effects of modifying the microenvironment on the growth and productivity of the cell or microorganism.
John Collins, Ph.D., senior applications scientist at NanoInk, will discuss the firm’s tip-based lithography. While the underlying Dip Pen Nanolithography® (DPN®) fabrication technology—based on microscale quill-like pens that can print features in the range of 1–10 microns—was developed about a decade ago, the company first began to use it for cell-based assay development in 2010.
Initial applications focused on the deposition of cells and extracellular matrix (ECM) proteins in precisely defined patterns on a surface to identify the binding sites of the proteins and to characterize their interactions with the cell membrane and the associated cellular responses.
NanoInk has expanded the technology to include arrays of dip pens that can simultaneously deposit multiple proteins or other biomaterials, including nanoparticles and polymer/drug complexes that can be internalized by cells, to produce arrays of cellular microenvironments that can be controlled and modified.
“The ability to precisely and simultaneously pattern different materials allows for the creation of single to a few microenvironment arrays,” explains Dr. Collins. One of several applications of the technology in development at NanoInk is the use of arrayed cellular microenvironments for in vitro toxicology studies during drug development.
The company recently demonstrated proof-of-concept of another application. A study illustrated the potential for single-cell co-culture by arraying fibroblasts and myoblasts within close proximity on a surface using direct deposition of the ECM proteins fibronectin and laminin in varied sizes and geometric organization. The results depicted differential binding of the two cell types to the proteins.

The Envirostat 2.0 allows cell type independent isolation, trapping, and microenvironmental controlled perfusion analysis of the single cell. An artificially colored HeLa cell, red blood cell, and bacterium trapped by negative dielectrophoresis (nDEP) in an octode electrode geometry adapted to cell dimensions are shown. The nDEP chip periphery on the right allows controlling temperature and chemical microenvironment surrounding the isolated floating single cell.
Tumor Cell Heterogeneity
Ola Söderberg, Ph.D., associate professor at Uppsala University, is studying the effects of post-translationally modified proteins and of protein interactions on signaling cascades in single cells.
By combining techniques that enable the simultaneous detection of nucleic acids and proteins in individual cells in fixed tissue sections, Dr. Söderberg can design multiplexed assays that have sufficient sensitivity to overcome some of the limitations associated with low signal-to-noise ratios or cross-reactivity that can compromise the usefulness of conventional FRET or single antibody-based assays, for example.
The approach developed by Dr. Söderberg combines two methods: a proximity ligation assay (PLA) and padlock probes. The PLA assay makes use of two antibodies, each of which recognizes and binds to a target protein molecule. Each antibody is linked to a short DNA strand, and when both antibodies are bound to the target, the strands combine to form a circular DNA molecule that generates a detectable signal.
Padlock probe technology allows for in situ detection of single mRNA molecules as a measure of gene expression. When used together, these methods can detect a gene transcript and its protein product in a single experiment.
Dr. Söderberg’s group is using this approach to measure multiple biomolecules simultaneously in a cell to identify signaling pathways that regulate gene expression. Specifically, their work focuses on cancer and characterizing the biochemical pathways that govern the growth and survival of cancer cells.
Ongoing studies aim to improve the efficiency of the assays and make them even more highly multiplexed, to use the assays for drug screening and for preclinical and clinical cancer research, and to explore the potential for developing assays for diagnostic purposes to characterize the heterogeneity of tumors, identify the differences between cell types, and understand how the various types of surrounding cells are affected by the presence of a tumor.
Anders Ståhlberg, Ph.D., a researcher at University of Gothenburg, Sweden, is performing gene-expression profiling at the single-cell level to identify the molecular mechanisms involved in sarcoma development and cancer stem cell differentiation. He describes the use of real-time reverse transcription quantitative PCR (RT-qPCR) to characterize cell types, biomarkers, cell differentiation, and cell functions with a level of sensitivity that allows for single-transcript detection.
Dr. Ståhlberg hopes that his research can help explain why a chemotherapeutic drug may kill 90% of tumor cells in a particular patient, allowing 10% to survive and proliferate. Are the surviving cells cancer stem cells—a concept not yet clearly defined—and how are they different?
Answering these types of questions requires single-cell resolution, according to Dr. Ståhlberg. With a better understanding of tumor cell heterogeneity, “we can then go back and monitor the system and treat the different populations of tumor cells,” he says.
An advantage of RT-PCR is that it is now commonplace in biological research. “Everyone knows what the data looks like and can do it at their own scale, starting at a small scale and building up,” says Dr. Ståhlberg. RT-PCR is easy to do and yields a great deal of knowledge, he adds. The main challenge in performing RT-PCR in single cells does not relate to the technique itself, but rather to sample preparation and, for example, extracting single cells from a tumor sample.
Each of the conventional methods for single-cell capture has benefits and drawbacks. With flow cytometry, for instance, you do not know where in the tumor a cell came from, and with microdissection followed by laser capture, the tissue samples have to be fixed before they can be analyzed, Dr. Ståhlberg explains.
Furthermore, the process of capturing single cells may alter their gene-expression profile and introduce bias. It is also difficult to know whether all of the different cell types in a tumor are represented in the sample being analyzed. Improved methods for sample prep upstream of RT-PCR are needed.

Fluorescent image of Fibronectin (red) and Laminin (green) demonstrating the ability to fabricate cell microenvironment arrays with various shapes of extracellular matrix proteins. [NanoInk]