The gene editing world has been revolutionized by the Nobel Prize–winning CRISPR-Cas9 system. Although CRISPR-Cas9 offers advantages over earlier genome editing methods, it isn’t necessarily the last word. Indeed, the past decade has revealed limitations of CRISPR-Cas9, which can’t tackle all the different types of disruptions in the DNA sequence underlying genetic disease. As there isn’t a one-size-fits-all solution for gene editing, there is enormous scope for biotech companies to mine nature to identify additional CRISPR nucleases and build on CRISPR to develop more flexible platforms for genome editing.
Prospecting for nucleases
Rather than passively wait for novelty, biotech company Metagenomi actively seeks it out. “Every quarter, we go out to numerous locations, and we take samples from the environment,” says Brian Thomas, PhD, the company’s founder and CEO. “It allows us to tap into life on this planet that we didn’t know existed and have never studied.”

Metagenomi
Thomas spent nearly 18 years in academic research at UC Berkeley. For 10 of those years, he worked with Jillian Banfield, PhD, a professor of earth and planetary science and a professor of environmental science, policy, and management. While in Banfield’s group, Thomas contributed to over 100 publications in the metagenomics/microbiome space.
More recently, this experience has been contributing to Metagenomi’s discovery platform technology and analytical expertise. By combing through metagenomic information, the company can identify novel cellular machinery. Then the company can adapt its finds into therapeutic tools.
Thomas argues that there are so many potential samples in the natural environment that Metagenomi can prioritize its sample collection based on certain features of organisms. For example, if the company were interested in CRISPR-like viral defense mechanisms, it could focus on getting natural samples from areas that are high in viral load. Doing so would, according to Thomas, improve Metagenomi’s chances of finding something similar to CRISPR.
But if one wants to acquire novel nucleases, one needn’t dig into privately sourced data derived from hard-won samples, that is, samples collected from the far corners of the world. Instead, one may use publicly available data sources, which are massive and fruitful. The less peripatetic approach is favored by Arbor Biotechnologies, a next-generation gene editing company founded by the Broad Institute’s Feng Zhang, PhD, the Wyss Institute’s David R. Walt, PhD, and two Zhang laboratory alumni, David Scott, PhD, and Winston Yan, MD, PhD.
“The advantage of the public data sets is that there’s just a lot of sequencing going around in the world,” says David Cheng, chief technology officer at Arbor Biotechnologies. “Lots of sequencing efforts end up deposited in public places that make it available for us to aggregate and self-annotate.”
Sifting for specific nuclease characteristics
A list of thousands of putative nucleases, whether from private or public data, is a good starting point, but it’s far from having an actual novel nuclease ready for human cells. To identify the most promising nucleases while parsing through massive amounts of genome sequence data, Metagenomi and Arbor Biotechnologies flag the nucleases that appear to have certain features.
One of the nuclease features prized by both companies is the ability to target the right spot in a DNA sequence. This feature depends on more than just the guide RNA. Adjacent to the target site, there has to be a corresponding protospacer adjacent motif (PAM)—a short DNA sequence (usually two to six base pairs in length) that follows the DNA region targeted for cleavage by a nuclease.
“The PAM sequence is part of the active structure of the enzyme,” Thomas explains. “If you don’t have that right active structure, the Cas9 won’t cut. So, different nucleases with specific PAMs are needed to account for the variety in sequence next to editing sites to target the right spot of DNA.”

Arbor Biotechnologies
In addition to targetability, an important characteristic is the nuclease cut type. “For example, if one is trying to do an excision, then having nucleases that cut in slightly different ways to have slightly different editing outcomes can result in greater variation,” Cheng notes. “So, that can give us [an opportunity] to pick the right nuclease for [a given] application.”
Off-target effects are another vital characteristic of putative nucleases. “One of the concerns about this novel technology is safety,” Thomas insists. “These things are going to be interacting with genomes, and we want to make sure they’re doing it in a controlled way.”
Since delivery is a key part of gene editing, Metagenomi and Arbor Biotechnologies have screened and prioritized tiny enzymes. “It frees up our ability to think about packaging limitations in the current systems of delivery,” Thomas relates. “But also, when you think about manufacturing, there are benefits. A longer molecule is going to take longer and cost more. And when you put a foreign protein into a mammalian cell, there’s a correlation between size and toxicity from the molecule itself.”

Arbor Biotechnologies
Regarding delivery modality, John Murphy, PhD, chief scientific officer at Arbor Biotechnologies, says that nucleases and nuclease guides can be delivered in the form of mRNA, and that the delivery vehicle can be a lipid nanoparticle. Nuclease delivery, then, can resemble the spike protein delivery achieved by the mRNA-based SARS-CoV-2 vaccines. However, alternative delivery methods may be preferable. As Murphy notes, with small nucleases, delivery systems such as adeno-associated viral vectors become available.
Turning up the base, anticipating prime time
Not everyone has to rifle through exotic nucleases to find gene editing success. Some are content to work with familiar nucleases, including good old CRISPR-Cas9. For example, Prime Medicine has modified CRISPR-Cas9 so that it nicks DNA rather than creating double-strand breaks in DNA. The “nickase” is fused to an engineered reverse transcriptase, resulting in a system that follows a search-and-replace approach and is capable of introducing point mutations. Essentially, the system offers an advanced form of base editing. It can introduce all 12 possible single-base substitutions.
Prime Medicine was founded by David R. Liu, PhD, and Andrew Anzalone, MD, PhD, who began developing their search-and-replace approach while they were still at Liu’s Broad Institute laboratory. At the end of September, Prime Medicine disclosed that it planned to go public. According to Renaissance Capital, Prime Medicine’s initial public offering could reach $200 million—a hefty sum for an initial public offering in a challenging market.

Beam Therapeutics
Other proponents of base editing include Beam Therapeutics. According to the company’s CEO, John Evans, base editing is the ultimate precision medicine. “Here we are making changes at the fundamental level of information in the genome, which is a single base,” Evans said at the State of Biotech, a virtual summit hosted by GEN Edge. “This is better than a small molecule. This is a platform.” He observed that base editing technology has already surpassed the feat of altering single bases. It is now possible to edit multiple sites at the same time, combining different types of edits to help treat a disease. He also suggested that multiplex editing could be more or less intense, allowing different medicines to be created that would present similar toxicology results, regulatory packages, and clinical characteristics.
Evans believes that with tools like base editing, the promise of helping as many people as possible has been unlocked. “Base editing represents one of the newer forms of editing,” Evans noted. “This is a sign of progress in the field. We’re continuing to evolve these tools.”
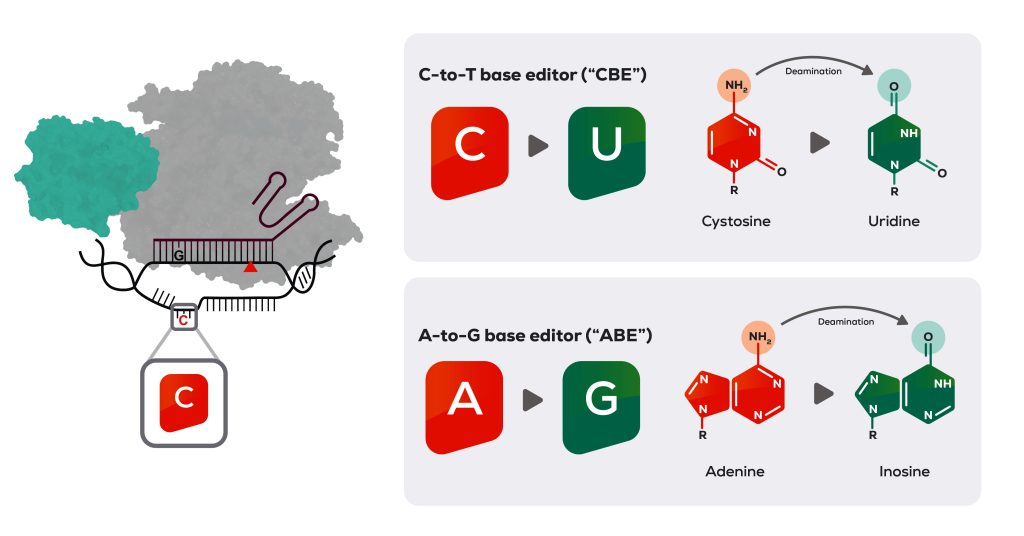
Another gene editing company that is working in the base editing space is PerkinElmer’s Horizon Discovery. Horizon Discovery is using exclusively licensed IP from Rutgers University for a novel base editor that’s part of the company’s Pin-point system.

PerkinElmer
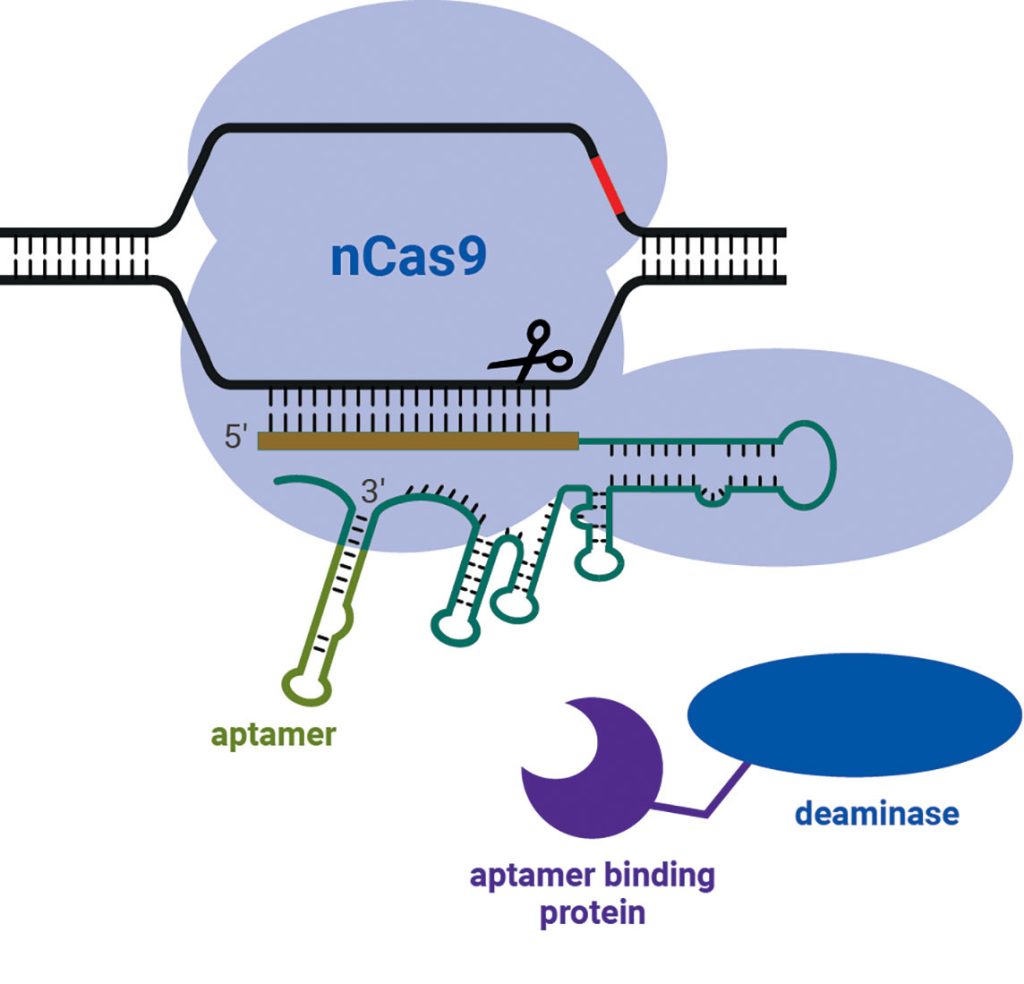
“It’s not the same as the other base editors that are on the market, and the main difference is that the Cas9 nickase is not fused to the deaminase required to make the nucleotide conversion,” says Michelle Fraser, business unit manager, base editing, PerkinElmer. “That’s important [because you now have] a modular system. So, you can optimize the chosen Cas enzymes and deaminases depending on where you want to edit.”
Together, the nickase and deaminase are recruited to target DNA by the guide RNA, which has an aptamer bound to it that matches an aptamer-binding domain on the deaminase. “Separating the nickase and deaminase allows us to do a knock-in and a knock-out all in the same edit in the same reaction,” Fraser points out. “We can do multiplex editing like the base editors can all do, but we can also knock in.” As a proof of concept, Horizon Discovery has introduced a CAR at the same time as knocking out all of the immunogenic markers on the T cell to make a CAR T cell.
Half of the equation
Nuclease tools are just one-half of the genome editing revolution launched by CRISPR. The other half has to do with CRISPR-associated effectors such as transposases.

Arbor Biotechnologies
“We view the nuclease as a key chassis in [the gene editing] toolbox,” says Devyn Smith, PhD, CEO, Arbor Biotechnologies. “You can then add or take away factors or whatever you’re trying to do with that nuclease to edit the DNA where you need make an edit.” Transposases, adds Arbor Bio’s Murphy, are “naturally evolved systems where the transpose system has co-opted a Cas to precisely insert a large cargo into DNA.”
At the end of the day, the CRISPR tool, whether a nuclease or an effector, will have to match the application or therapeutic. Each genetically defined disease (and possibly each distinct cohort of patients with that disease) may call for the development of customized CRISPR systems as therapeutics. To answer the call, we must first have the entire toolkit in front of us.