January 1, 2012 (Vol. 32, No. 1)
Finding Applications in the Improvement of Protein Production and Processing Operations
Many recombinant therapeutic proteins such as monoclonal antibodies are produced in eukaryotic cells to enable post-translational modifications such as glycosylation. Glycan profiles and charge variants are important quality attributes for the activity, efficacy, and safety of therapeutic proteins.
To meet these parameters highly efficient and targeted media development is crucial. Whereas systematic approaches such as QbD are increasingly applied, a deeper understanding of the interaction between metabolic alterations, protein production, and processing should significantly improve production processes (Figure 1).
Metabolomics is a valuable tool to study underlying mechanisms and involved biochemical pathways—especially for the widely used CHO cell line, for which little is known about its physiology.
In this tutorial we will show how we evaluated the potential of metabolomic tools to support cell-line characterization and subsequent process development, this information was also recently presented at the ESACT meeting in Vienna. As part of the study, we compared a CHO producer and its corresponding mock cell line (transfected with an empty plasmid) on a metabolic level in order to see whether the metabolite profile differences came from the increased protein production capacity. In addition, we were able to observe the complex glycosylation machinery.

Figure 1. Cell culture cGMP manufacturing at Sandoz in Schaftenau, Austria
Sample-Prep Optimization
First, studies dealing with optimization of sample preparation—more precisely the sampling procedure called quenching—were conducted. This step is responsible for the mandatory immediate stopping of enzymatical reactions in order to preserve the intracellular metabolite environment and can strongly affect subsequent results.
We developed a simple and straightforward protocol, with no need for special equipment, that focuses on the efficiency of stopping metabolic activities and the potential metabolic leakage due to the loss in membrane integrity.
Within this protocol, it is recommended that cells are quenched and washed simultaneously to keep sampling time to a minimum and to prevent further metabolic activity within the cells. Studies also showed that by using this procedure additional washing steps can be omitted. A detailed description of the studies will be published elsewhere. An overview of the key steps of the implemented sampling procedure, which allows simultaneous quenching of a large number of samples, is illustrated in Figure 2.

Figure 2. Overview of the key steps for adequate quenching of mammalian cells in suspension
Targeted Metabolomics Methods
Targeted metabolomic analyses were performed for cell culture supernatants as well as for cell lysates. For extraction of intracellular metabolites freeze/thaw cycles after resuspension in phosphate buffer were used.
Commercially available AbsoluteIDQ KIT plates were used for the quantification of amino acids, hexose, and biogenic amines. This fully automated assay is based on PITC (phenylisothiocyanate) derivatization in the presence of internal standards followed by LC-MS/MS detection using a mass spectrometer with electrospray ionization.
The quantitative analysis of energy metabolism intermediates (glycolysis, citrate cycle, urea cycle) was accomplished with a hydrophilic interaction liquid chromatography (HILIC)-ESI-MS/MS method used in highly selective negative MRM detection mode.
Intracellular amounts of nucleotides and nucleotide sugars were analyzed by liquid chromatography-electrospray ionization mass spectrometry on surface-conditioned porous graphitic carbon.
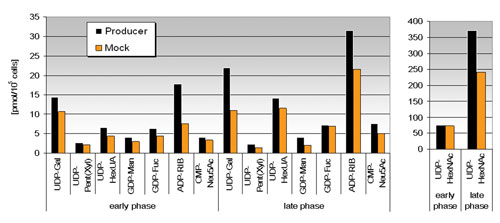
Figure 3. Intracellular nucleotide sugar concentrations: Producer cells showed increased nucleotide sugar concentrations, which reflected the enlarged demand on glycosylation processing compared to the mock cell line. Early phase represents data within exponential phase (up to and including day 7), whereas late phase shows averaged data of day 10 and 12.
Producer vs. Mock Cell Line
Both CHO cell lines were cultivated as a fed-batch process lasting for 14 days in serum-free, chemically defined in-house medium in a 15 L bioreactor. Sampling for subsequent metabolite quantification took place at various time points throughout cultivation.
In addition to the cell-line comparison, it was possible to characterize different growth phases during the standard production process. Major metabolic distinctions were observable between the exponential phase (up to and including day 7) and the later phase compromising stationary and apoptotic phases for both cell lines.
To compare producer and mock cell-line cultivations, multivariate data analysis using a PLS model on all quantified compounds was employed. Changes between different sampling points, which represent the different stages of cultivation, showed stronger influence on the prediction model than overall alterations between the tested cell lines.
Therefore, differences between tested cell lines have to be evaluated for each phase individually as no overall variation was detectable.
Major variance between producer and mock cell lines within the individual cultivation phases was detected for intracellular nucleotide sugar concentrations, as shown in Figure 3.
Nucleotide sugars play an important key role in the mammalian glycosylation pathway. Increased glycosylation processing in producer cells seemed to have led to higher intracellular concentrations of required precursors for glycan structures due to increased stimulation of the overall glycosylation machinery.
Alternatively, increased consumption by producer cells should have led to lower concentrations or even to limiting bottlenecks. Increased amounts of intracellular nucleotide sugars at late stage for both cell lines were evident, but an adequate explanation with respect to cell and process knowledge was not found.
Moreover, specific consumption/production rates of diverse metabolites were different between producer and mock cell lines within the early phase (exponential phase comprising sampling between day 3–7). Mostly, amino acids were affected. In general, mock cells showed a slight increase in energy demand reflected by higher specific consumption rates. Thus, especially the slightly higher growth rate of mock cells seemed to be of relevance resulting in differentiation between proliferating and producing cells.
Conclusion
In this study, we detected expected differences between a producer and its corresponding mock cell line and thus we were able to prove the applicability of intracellular quantification of metabolites for bioprocess development.
Metabolomic analyses might also detect variations between different host cell lines encoding for the same recombinant protein or metabolic differences due to modified process settings. Potential biomarkers could be identified and used for systematic cell line and clone selection.
Furthermore, intracellular metabolite quantification might identify bottlenecks in protein production or processing that are not noticeable with current standardized monitoring and spent media analysis.
This expertise will be used for further improvement of our process-development strategies in order to deliver efficient and robust processes yielding high amounts of recombinant proteins with the desired product quality.
Jennifer Kronthaler ([email protected]) is a researcher, and Christine Heel, Ph.D., is group head, cell biology and medium development at Sandoz Biopharmaceuticals.



