April 15, 2017 (Vol. 37, No. 8)
MALDI-FTMS Emerges as a Valuable Tool in Small-Molecule R&D
The situation in small-molecule pharmaceutical R&D has been well described in recent years—low numbers of new drugs approved and the rising cost of bringing a compound through clinical development to market. Against this unrelenting pressure on discovery and development, one mantra that has stuck is “fail early, fail cheap.”
This is perhaps not surprising, given that many drugs fail due to unexpected results late in clinical trials. Pharma companies are increasingly aware of the disconnection between outcomes in clinical programs and early-stage development work. As part of the industry’s response, ADME/TOX (absorption, distribution, metabolism, excretion/toxicity) investigations are now integrated as early as possible in the pathway, in order to understand essential details of drug distribution, metabolism, and toxicology ahead of going into clinical trials.
Current Tools
Traditional approaches and tools available for the risk assessment of safe and efficacious drugs have been based on measuring the parent drug in plasma—both in animal models and humans. This often does not provide a sufficient level of information to the scientist. It is recognized that most drug targets are not in plasma, and determining the relevant tissue distribution of not only the parent drug but also its metabolites would provide much greater understanding of pharmacology and toxicology.
The current best practice methods of quantitative whole body autoradiography (QWBA) and liquid chromatography-mass spectrometry (LC-MS) put the emphasis on tissues rather than plasma, but still fall short of providing the complete picture. Both techniques have challenges. QWBA is the technique of choice for determining drug distribution,1 and the data generated is often primarily provided in new drug regulatory submissions. The technique, however, presents a composite of the total radioactivity present and cannot distinguish parent drug from metabolite. Thus it has severe limitations for researchers looking for insight into biochemical pathways and mechanisms.
LC-MS analysis is performed on extracts from tissue homogenates. This type of analysis does not elucidate any spatial information and, just as importantly, can be misleading. For example, if an analyte in the tissue is highly localized, the homogenization process could potentially dilute it below the limit of detection (LOD). Alternatively, when an analyte is determined from tissue homogenate, a researcher could draw incorrect conclusions about toxicity, because the analyte is presumed to be evenly present throughout the tissue when in fact it may be highly localized.
New Thinking Required
Over the past five to seven years, a new approach, taken from preexisting technology, that provides quantitative, spatially resolved data on the tissue distribution of drugs and drug metabolites has emerged. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) is uniquely suited to this task. Figure 1 shows how the MALDI MSI experiment is initiated by mounting a tissue section onto a target, applying a matrix and rastering a laser across the surface of the tissue. At each discrete location where the laser is fired, an accurate mass spectrum is acquired. By plotting the ion intensities as a function of the x and y coordinates on the tissue, ion images are generated. Software co-registers tissue histology images with these, allowing a researcher to visualize biology and chemistry in an integrated fashion.
A range of mass spectrometers can be integrated into a MALDI MSI system, depending on application needs. MALDI-TOF systems, for example, offer high throughput (with lower ability to distinguish molecules of similar molecular weight) while Fourier transform mass spectrometers (MALDI-FTMS) provide the ultimate in measurement accuracy and mass resolving power. High MS performance features of MALDI-FTMS can often yield unique molecular formula identification for ion images.
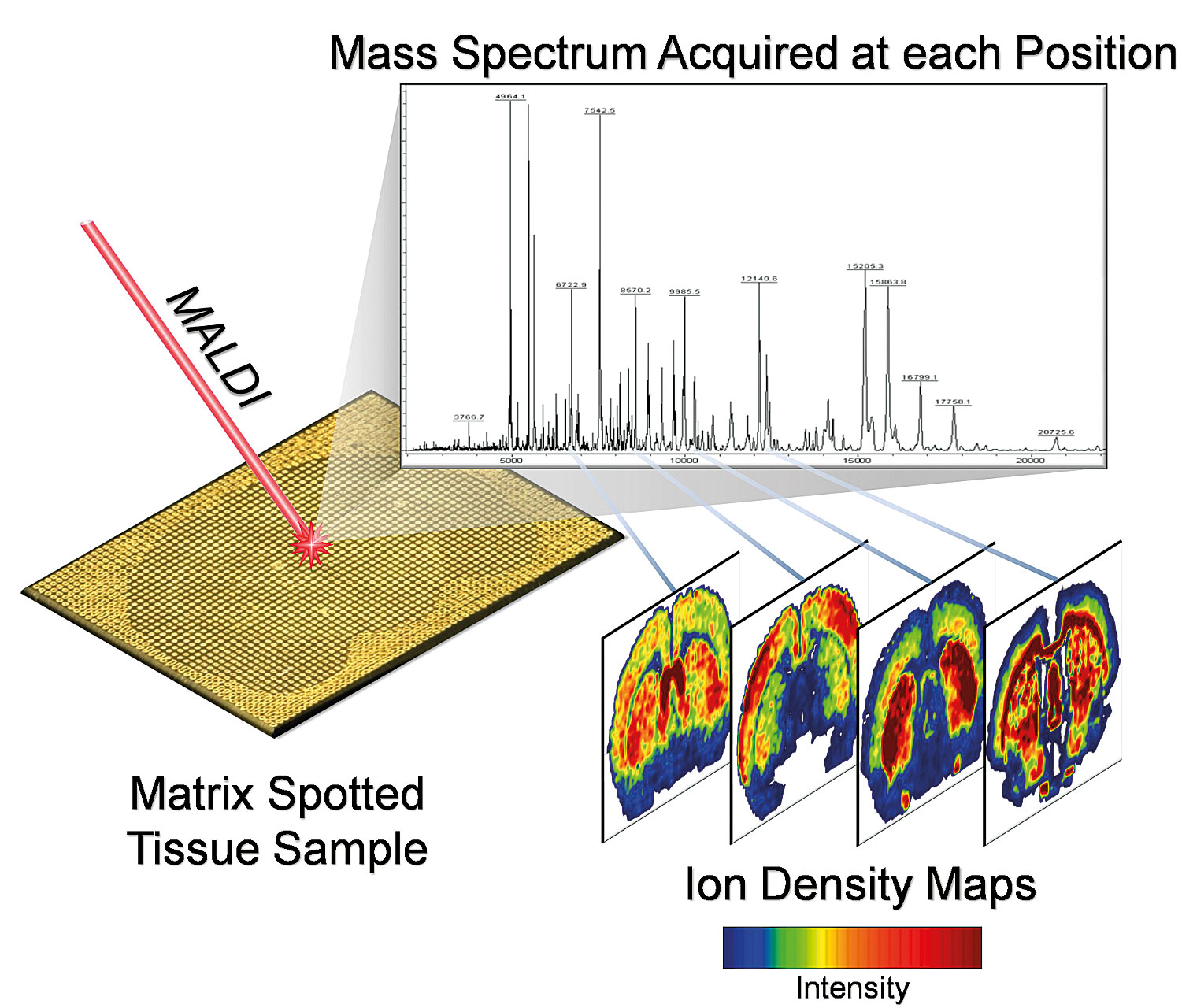
Figure 1. A MALDI MSI experimental protocol
Principles and Practice
MALDI MSI takes a conventional tissue section and coats it with a fine aerosol of MALDI matrix solution, which extracts molecules from the tissue, but retains the spatial relationships found in the underlying tissue. Following preparation, the sample is measured in the mass spectrometer. The result is spatially resolved, accurate mass spectra. Because the laser only probes the matrix crystals that are on the surface of the section, the underlying cellular features are not disrupted and can be taken through a standard histological staining routine, so that a high-quality histology image can be captured for co-registration.
By merging histology with the molecular information from the MALDI mass spectrometer, a histology-directed analysis of the tissue is possible. Figure 2 highlights an extract of early work by S. Castellino et al.2 A small area of a histology section is magnified showing regions of inflammation. An ion map (50 µm spatial resolution), which corresponds to the same zoomed region, indicates the localization of lapatinib metabolite, M10, is only in the regions associated with the inflammation.
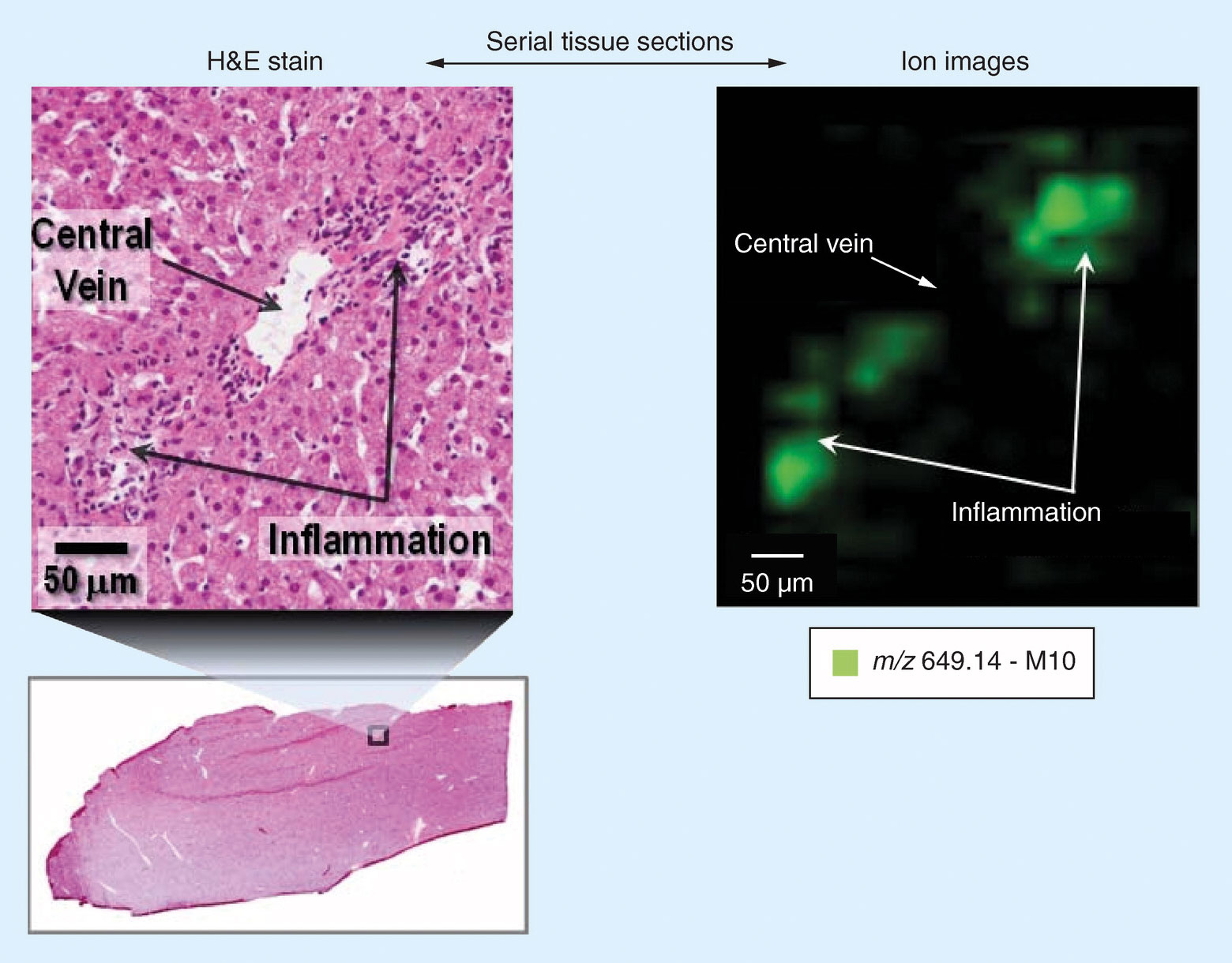
Figure 2. Correlation of histology and MALDI MSI image in dog liver sections
A recent publication by M. Reid Groseclose et al.3 evaluated the additional information that MALDI MSI could provide over and above LC-MS in a nephrotoxicity study on the anticancer drug Dabrafenib (DAB) in rats. This work was in support of developing DAB for use in pediatric patients. Previous studies had identified some unexpected, adverse kidney effects, including tubular deposits in some juvenile rats. In this study, MALDI MSI was used to determine the distribution of DAB and primary metabolites in the kidney. Subsequent MSI analysis of the tubular deposits revealed them to be calcium phosphate crystals.
Many other papers, posters, and presentations have highlighted the power of these new combinations. For example, work on Chloroquine in a Long-Evans male rat model by G. Hochart et al.4 using a MALDI-FTMS system (Bruker SolariX XR, Figure 3), reported that: “Combining the strengths of QWBA and MALDI MSI allowed for the sensitive detection of drug related material (QWBA) and the quantitative differentiation of parent and metabolite (MALDI MSI).”

Figure 3. Bruker SolariX XR
Challenges and Opportunities
MALDI MSI using FTMS is still a relatively young technique. Advances in instrumentation and sample processing that have occurred over the past five to seven years have shown the value of the technique to the pharmaceutical industry, where it has been included as supplementary data to regulatory filings. As the industry continues to recognize the value of MALDI MSI to provide additional insight and inform decisions regarding the development of a candidate compound, we are sure to see such data become more common in regulatory submissions and eventually become required.
It is easy to imagine that the combination of MALDI MSI with QWBA and LC-MS will yield deeper understanding about a drug, and its pharmacology, and toxicology, to deliver tangible benefits as it moves through development to market.
Dale Shannon Cornett, Ph.D. ([email protected]), is applications development manager, mass spectrometry, at Bruker Daltonics.
References
1. Pharmaco-Imaging in Drug Biologics Development: “Fundamentals and Applications” pg171, Brian R. Moyer, Narayan P.S. Cheruvu, Tom C.-C. Hu, Editors
2. Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011 Nov; 3(21):2427-41
3. M. Reid Groseclose, Susan B. Laffan, Kendall S. Frazier, Angela Hughes-Earle, Stephen Castellino. Imaging MS in Toxicology: An In-vestigation of Juvenile Rat Nephrotoxicity Associated with Dabrafenib Administration. Journal of The American Society for Mass Spec-trometry., 2015, 26(6):887 – 898
4. G. Hochart et al, Comparative Approaches between Quantitative-MSI and Quantitative-WBA: Application to Chloroquine Administra-tion in a Long-Evans Male Rate Model, Presented at ISSX conference, (Accessed March 2017)


