In The Hunger Games, Katniss Everdeen shocked the distracted gamemakers seated above her by shooting an arrow and blasting an apple out of the roasted pig’s mouth on the buffet table. Accuracy counts!
Pharmaceutical and biotech firms are waging their own hunger games, albeit a bit less dramatic, as they attempt to hit the bullseye of identifying and validating therapeutic targets. New approaches are sorely needed as during the last several decades, drug discovery pipelines have suffered from high attrition rates for candidates entering clinical trials.
The recently held Cambridge Healthtech Institute’s 15th Annual Target Identification and Validation conference focused on innovative chemical biology and phenotypic screening advances to overcome the challenges of pinpointing novel, druggable targets. Solutions discussed included a zebrafish platform for high-throughput screening, an innovative high-throughput genome engineering strategy to identify essential genes for cancer immunotherapy, a high-content autophagy screen to discover pathways in disease biology, and defining intracellular pathways that mediate lung fibrosis.
Zebrafish-based target validation platform
Approximately 95% of compounds fail after Phases II and III, highlighting the need for fishing out innovative strategies to streamline the drug discovery pipeline.
Zebrafish may be swimming to the rescue.
“To date, the majority of animal testing is performed in mammals, making the whole process laborious, costly, and time-consuming,” declares Vincenzo
Di Donato, PhD, project manager of the genome editing platform at ZeClinics. He adds, “We overcome these limitations by exploiting the advantages of zebrafish, a valuable alternative model for preliminary phases of drug screening pipelines.”
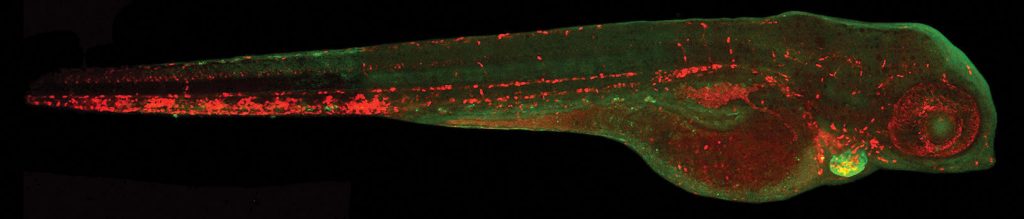
in the erythrocytes.
Di Donato says that the high genetic and physiological homology between humans and fish enable a detailed study of the function of potential therapeutic targets as well as a deeper understanding of the mechanism of action of small molecules on those targets.
“The elevated reproduction rate and fast organ development of zebrafish allow for high-throughput screenings resulting in a great time reduction,” he explains. “Low maintenance costs of fish colonies and reduced quantity of molecules required for assays have an enormous impact on the investment required for drug development. Importantly, drug screening on zebrafish larvae is performed at early developmental stages, before 120 hours post-fertilization, a time when they start being subject to regulations for animal experimentation.”
The company’s ZeCardio® screening platform provides a tool for genetic association studies in cardiovascular disease. Di Donato, elaborates, “This methodology combines the advantages of the CRISPR- Cas9 system for generating somatic mutant larvae by using guide RNAs targeting candidate genes with high mutagenesis efficiency (up to 100%) and a high-throughput in vivo imaging system allowing for morphological and functional analysis of cardiovascular phenotypes.”
Some therapeutic molecules identified in zebrafish-based screenings have already progressed to the clinical phase (e.g., PDE5A inhibitors for the treatment of Duchenne muscular dystrophy). However, challenges remain that Di Donato believes are solvable. He reveals, “To further improve the translatability of zebrafish in drug discovery, ZeClinics is actively working on the development of humanized zebrafish models.”
Genome-wide net for cancer genes
Most cancers spawn genetic aberrations as a product of their neoplastic evolution. Sometimes somatic mutations can generate antigens capable of eliciting T-cell responses, yet some also induce resistance to T cell–based immunotherapies.
“Cancer is a disease of the genome,” emphasizes Neville E. Sanjana, PhD, assistant professor, departments of biology, neuroscience, and physiology, New York University. “Tumors can harbor up to one million mutations. This presents major difficulties in understanding and identifying which are central to the disease and which are passengers.”
Sanjana is also pursuing another lofty goal—the complete genome. He emphasizes, “The region outside of genes is an even larger landscape. Less than 2% of the human genome consists of protein coding sequences. Most of the 3 billion other bases consist of noncoding areas that likely serve key regulatory roles.”
Although seemingly akin to finding the proverbial needle in the haystack, Sanjana and colleagues have already identified mutations in promoter genes that increase drug resistance. They will next interrogate the sequence around genes vital to immunotherapy success.
He concludes, “The noncoding genome represents a fascinating arena for developing and conducting large-scale gene assays to identify regions important for modulating immunotherapies. We have only begun to understand the functional importance of specific tumor mutations in coding and noncoding regions. My hope is that such an understanding will ultimately lead to more precisely tailoring treatments against cancer.”
Autophagy modulators
Autophagy, literally meaning “eating of self,” is a critical homeostatic mechanism for cellular turnover. But there are two sides to this hunger game: activation and inhibition. Both can be used as therapeutic targets. For example, some cancer cells use autophagy as a survival mechanism. Thus, the identification of autophagy inhibitors could help attack tumors.
On the activation side, Stephen Walker, senior research scientist III, molecular
characterization and screening, AbbVie, comments, “Our area of focus is on the
identification of autophagy activators as therapeutic targets since these may help clear and degrade protein aggregates
associated with neurodegenerative diseases such as Huntington’s, Parkinson’s, and
Alzheimer’s disease.”
Walker and colleagues are using a multiparametric high-content screening approach utilizing an annotated chemical library (ACL) consisting of approximately 25,000 compounds to identify targets and pathways that induce autophagic endpoints in neuronal cell models.
“This is quite different than our traditional high-throughput screening approach where approximately 900,000 compounds are typically tested in biochemical, biophysical, or cell-based assays with the goal of identifying well-characterized compounds for initiating medicinal chemistry efforts. Our goal from the autophagy screen is to identify hits and discover the targets and pathways involved in the disease biology,” he explains.
Prior to the initial screen, Walker characterized and validated multiple endpoints of autophagy using a well-annotated compound collection of approximately 1500 FDA-approved drugs. Endpoint indicators included markers of transcriptional activation, phagophore formation, and autophagic flux.
“Using this approach, we identified many known regulators of autophagy,” he reports. “We are currently using this panel of endpoints as part of a larger strategy to deconvolute hits from neuroscience-specific screening endpoints using the ACL and to evaluate whether the observed phenotypes have an autophagic mechanism of action.”
These studies have already identified several promising hits belonging to distinct target classes that modulate the desired phenotype through autophagy. According to Walker, “Some of these targets are novel to the neuroscience pathways that we are studying, but have known implications in other diseases. We are hoping to leverage previous knowledge and utilize potent, specific chemi-cal matter to help with target validation and to speed the drug discovery effort of these targets for new indications in neuroscience.”
Targeting lung fibrosis
To nonscientists, the word “Yap” could signify a dog barking, a person droning on, or an island in Micronesia. But to scientists, YAP, or Yes-associated protein, is a potent transcription coactivator of genes involved in cell proliferation (as an activator) and in apoptosis (as a suppressor).
Harvard Medical School.
Santos is studying idiopathic pulmonary fibrosis (IPF), a lethal, progressive disease driven by pathological fibroblast accumulation and differentiation. IPF results in stiffening, thickening, and scarring of lung tissue, ultimately causing life-threatening complications such as respiratory failure. Currently, this fatal disease only has limited therapeutic options.
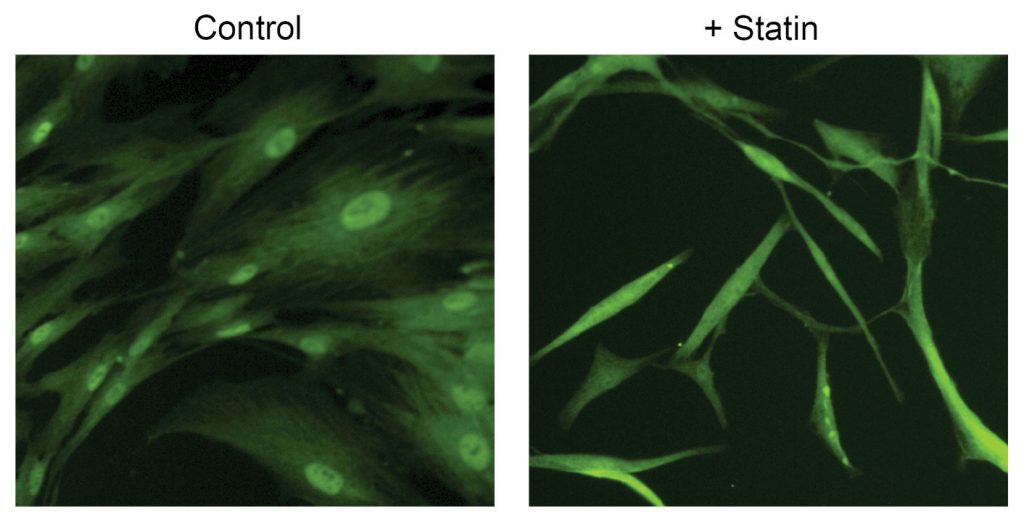
Working with colleagues in collaboration with Boehringer Ingelheim, Santos designed a small-molecule high-throughput screen in primary human fibroblasts to identify and validate YAP inhibitors as well as new pathways controlling YAP activation. The scientists utilized a high-content imaging screening method that not only allows imaging of individual cells but also identifies YAP cellular localization. She elaborates, “Since YAP is active in the nucleus, we sought compounds that would cause it to translocate out of the nucleus.”
Surprisingly, the group identified multiple human statins, i.e., HMG-CoA reductase inhibitors, which promoted YAP nuclear egress. “We validated these results by using the identified hit compounds to perform in vivo studies in mice,” she reports. “Evaluation of imaging flow cytometry data revealed statins are able to modulate YAP in mice lung fibroblasts. Importantly, statin treatment reduced established fibrosis in mice.”
Going forward, Santos plans to publish the results and to next develop a siRNA screen of the druggable genome for other YAP inhibitors. “We will continue to look for novel targets and seek proof of concept. We hope that our studies may ultimately lead to the development of antifibrotic therapies.”
As scientists develop new tools and strategies, the hope remains that these efforts will uncover validated targets, eventually leading to improved and safer therapeutics.


