March 1, 2018 (Vol. 38, No. 5)
A Molecular Interaction Detection Platform Uses a Gel-based Readout
It may not be long before nanoscale devices that function with the flip of a DNA nanoswitch are available in the clinical setting.
They could be used to easily and cost-effectively detect very low levels of biomarkers for diagnostic purposes, or to deliver therapeutic cargo when prompted to do so by a change in pH or recognition and binding of a target ligand, for example.
Wesley Wong, Ph.D., from Harvard Medical School, Boston Children’s Hospital, and the Wyss Institute for Biologically Inspired Engineering at Harvard University, and Ken Halvorsen, Ph.D., from SUNY Albany, jointly developed programmable DNA-based nanoswitches in Dr. Wong’s lab at the Rowland Institute at Harvard. These nanoswitches are based on emerging research on DNA origami (Figure 1).
Using the single-stranded genomic DNA of the M13 bacteriophage and mixing it with predefined synthetic oligonucleotides, the researchers were able to use hybridization as a way to place molecules (e.g., a receptor and a ligand, or pairs of antibodies) in prescribed locations along the genomic DNA strand. The resulting DNA nanoswitch exists as a linear duplex in the “off” state and a looped construct in the “on” state, with the predetermined conformational change being induced by binding events between molecules tethered to the DNA scaffold.1 The binding event essentially flips the switch.
Dr. Halvorsen and colleagues at The RNA Institute at SUNY Albany have created DNA nanoswitches that can detect microRNAs (miRNAs), which play an important role in gene regulation.2 “We can now use these nanoswitches to detect one specific miRNA out of a total cellular extract or tissue source in as little as 1 hour,” he says. This can be achieved without the need for amplification, labeling, use of enzymes (and their storage requirements), or the complex equipment that existing RNA detection methods typically require.
The readout for miRNA assays based on this nanoswitch relies on gel electrophoresis, taking advantage of the huge mobility shift between a linear duplex and a loop construct. While gel electrophoresis is a ubiquitous research tool, a more hands-off, automatable readout might be needed for the technology to translate to the clinic. “In my opinion,” says Dr. Halvorsen, “there really hasn’t been an effort to engineer gel electrophoresis to make it compatible with the clinic. The question of readout is a valid one. For the next stage, we will either have to reimagine how we run the gels, or we will have to translate the technology without a gel.”
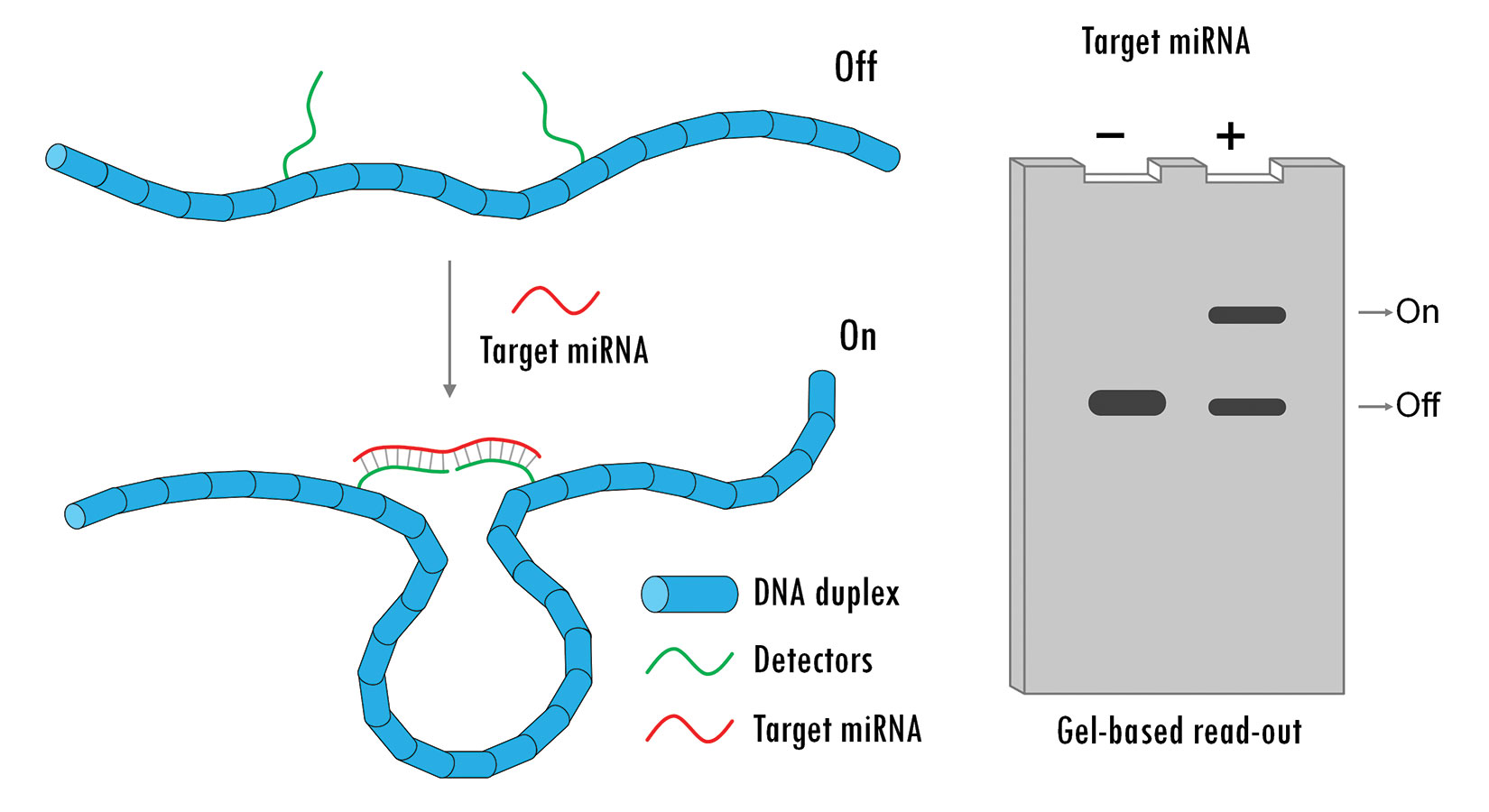
Figure 1. A DNA nanoswitch is designed to change conformations in response to a specific microRNA target. This conformational change can be easily detected on a standard agarose gel. [Ken Halvorsen, University of Albany]
A Universal Detection Platform
In addition to being programmable and modular, DNA nanoswitches offer advantages such as the ability to detect multiple different target molecules (e.g., proteins and nucleic acids) simultaneously. “The DNA scaffold serves as a universal detection platform,” says Dr. Wong. In addition, Drs. Halvorsen and Wong have shown that they can detect and characterize not just two-body interactions, but also four-body interactions. Different band patterns will appear on the gel depending on how many binding events take place.
Dr. Wong’s group at Harvard has used the DNA nanoswitch technology to develop the nanoswitch-linked immunosorbent assay (NLISA), as described in PNAS.3 Compared with the standard ELISA, NLISA is faster, less costly, and easier to run, in general. It is also more sensitive and less specific than conventional ELISA. NLISA is a surface-free technique, so there is nothing for proteins and other biomarkers to stick to and cause false-positive results. Reduced cross-reactivity as a result of a kinetic proofreading purification step also helps to reduce background noise. Dr. Wong envisions clinical applications for NLISA at the point-of-care for infectious disease detection, for example, or quantification of cardiac and other disease-related biomarkers for rapid diagnosis.
“We have been pushing the sensitivity of the assay in the lab and have improved it by multiple orders of magnitude compared to our previously published work, which was at the femtomolar level,” says Dr. Wong. The researchers are also developing multiplexing capabilities to enable simultaneous detection of multiple biomarkers in parallel, and are considering how this technology could be integrated into a user friendly, handheld device.
Measuring Antibodies in Clinical Samples
The simplicity, versatility, and ease of engineering of synthetic DNA are the main attributes that make it an ideal molecular substrate for creating nanoswitches, according to Francesco Ricci, Ph.D., and Alessandro Porchetta, Ph.D., from University of Rome Tor Vergata, Italy, and co-lead authors of a paper in Chemical Science that describes the allosteric DNA nanoswitches they designed for controlled release of a molecular cargo by biological inputs.4
Synthetic DNA is affordable to produce, according to Dr. Ricci, and “we can attach different molecules to it, such as molecules that provide a measurable output signal (optical or electrochemical), or recognition elements that can target various analytes (e.g., antibodies and other proteins). Also—and this is very important for us—DNA has a very simple and predictable chemistry, which means that we are able to design DNA strands that can adopt different conformations or that can switch between different states in a very straightforward way.”
Dr. Ricci’s group is developing DNA-based nanoswitches and nanodevices for clinical applications such as diagnostics (recognition of disease biomarkers) and drug delivery. For diagnostic use, the nanoswitch changes conformation when a specific biomarker binds to its recognition element, resulting in a measurable change in signal output. The nanoswitches used for drug delivery are designed to release a therapeutic cargo on biomarker binding.
“We are collaborating with a start-up company to commercialize a DNA-based nanoswitch that is able to measure different antibodies in clinical samples,” says Dr. Ricci. This can have important implications as point-of-care devices for rapid and low-cost diagnosis of infectious diseases. A striking advantage of our platform is its versatility; simply changing the recognition element on the DNA nanoswitch will make it possible to detect different antibodies.”
In addition to developing DNA-based nanoswitches that function through a conformational change mechanism, the researchers are adopting new strategies. These approaches mimic the function of biomolecular receptors “to respond in an optimal way to a specific target in the very complex environment of a cell,” notes Dr. Ricci. “Some of these ‘tricks,’ like allostery and cooperativity, allow us to control and tune the response of a receptor in a very precise way.”
Another aspect of the researchers’ work focuses on the development of triple helix DNA-based switches. DNA triplexes can form only at certain acidic pH levels, allowing for the function of triple helix-based switches to be regulated by changing the pH of the solution. “As pH dysregulation in the cell is important in some diseases,” explains Dr. Ricci, “we envision that these types of pH-dependent nanodevices will be useful for better understanding the mechanisms of these diseases or for developing tools that can release a therapeutic cargo only under certain pH conditions.”
It is also possible to design DNA strands that bind a target DNA to form a triplex through a clamp-like mechanism. This provides a novel form of conformation change with the potential for new applications in drug delivery. The researchers reported on clamp-like triplex nanoswitches that can be designed as nano-slingshots to “shoot” a drug only in the presence of specific marker antibodies (Figure 2).5

Figure 2. A nanoscale molecular slingshot made of synthetic DNA that is 20,000 times smaller than a human hair. This molecular slingshot can “shoot” and deliver drugs at precise locations in the human body once triggered by specific disease markers (i.e., antibodies). [Marco Tripoli]
Stress-Sensitive Hydrogels
Hans Heus, Ph.D., and colleagues at The Netherlands-based Institute for Molecules and Materials, Radboud University, have developed biomimetic stress-sensitive hydrogels controlled by DNA nanoswitches.6 The researchers sought to create synthetic polymer-based hydrogels that replicate the chemical and physical properties of natural hydrogels, and in particular, the ability of the gels to become stiffer when deformed (or when placed under stress). This is a crucial behavior for cells, which helps them prevent large deformations and protects them from rupturing when highly stressed. This ability for cells—altering their own rigidity based on environmental conditions—is believed to play an important role in cell differentiation and migration.
Dr. Heus’s group developed a synthetic hydrogel comprised of networks of bundled polymers functionalized with oligo tails with the capacity to stress-stiffen. The group then expanded the controllability of this system by incorporating functional DNA elements, or crosslinkers based on DNA nanoswitches. The elastic properties of the hydrogel are determined by the molecular structure of the crosslinker, resulting in a DNA-responsive hydrogel with tunable stiffness. The researchers designed the DNA nanoswitches to be responsive to pH (an i-motif and triple helix) or to ligand binding (a thrombin aptamer), giving them control over hydrogel stiffness (reversible change between tight and relaxed states) and, particularly, the onset of stress stiffening, which they have shown to have a role in controlling stem cell fate in 3D biomimetic scaffolds.
1. M.A. Koussa et al., “DNA Nanoswitches: A Quantitative Platform for Gel-Based Biomolecular Interaction Analysis,” Nat. Methods 12, 123–126 (2015).
2. A.R. Chandrasekaran, J. Zavala, and K. Halvorsen, “Programmable DNA Nanoswitches for Detection of Nucleic Acid Sequences,” ACS Sens. 1(2), 120–123 (2016).
3. C.H. Hansen et al., “Nanoswitch-Linked Immunosorbent Assay (NLISA) for Fast, Sensitive, and Specific Protein Detection,” PNAS 114 (39), 10367–10372 (September 2017).
4. M. Rossetti et al., “Allosteric DNA Nanoswitches for Controlled Release of a Molecular Cargo Triggered by Biological Inputs,” Chem. Sci. 8, 914–920 (2017).
5. S. Ranallo et al., “Antibody-Powered Nucleic Acid Release Using a DNA-Based Nanomachine,” Nat. Comm. 8, 15150 (2017).
6. A. Idili, A. Vallée-Bélisle, and F. Ricci, “Programmable pH-Triggered DNA Nanoswitches,” J. Am. Chem. Soc.136(16), 5836–5839 (April 9, 2014).


