
Genome engineering is advancing at lightning speed, but we want it to move even faster. Investment capital is surging into platform technologies now that the pharmaceutical industry has seen how quickly the biotech sector has developed COVID-19 vaccines and diagnostics.
It has become apparent that new technological solutions are arriving faster than development cycles can turn, potentially raising new commercial risks, particularly in manufacturing. But nobody is calling for a more deliberative pace. Everybody wants to race ahead.
The result? New solutions in biomedical research are propelling the field by leaps and bounds ahead of what was accomplished in decades past. Biotechnology and particularly genome engineering are driving new paradigms for healthcare access and contributing to a new healthcare ecosystem.
These trends prompted Synthego to define its core mission: the application of engineering principles in life sciences development to make gene therapies accessible. In one dimension, this focuses on solving complex gene editing problems while constantly engineering safely. In another, the focus is on engineering complex disease models to build corrective gene therapies using sophisticated screening and machine learning.
Looking to 2021 and beyond, we see that a few topics in the genome engineering space are worthy of discussion. Each of these topics amounts to a technological tipping point: ensuring the safety of gene editing, increasing the accuracy of discovery screening, and laying the foundation for a regulatory path for CRISPR-based gene therapies.
Improving safety with on/off-target analytics
One of the challenges in the ex vivo cell and gene therapy space, and particularly with induced pluripotent stem cells (iPSCs), is interpretation and risk mitigation of allelic change. Many gene variants that raise the risk of ectopic expression or cancer form a test profile against which sponsors must defend safety and risk profiles. The selective pressures that single-cell cloning or bioreactor expansion parameters put on cell populations generates disqualifying data at high frequency. While many of these issues are pinned to cell culture methods, a similar background occurs with genome engineering related to weak off-target homologies relative to the guide sequences targeting the primary locus.
It is possible to address that challenge by using automated workflows and engineering the process. By developing a statistical basis for understanding what went well and what went sideways, machine learning can support highly efficient gene engineering while quantifying off-target sites. Showing that instances of off-target editing are predictable and quantifiable contributes to risk management. As regulatory dialogue raises the importance of showing targeting profiles from hundreds of runs to quantify off-target risk frequency, we can build a rapid lot release assay based on a qualified gene editing process that not only improves risk management, but brings down costs.
At Synthego, we view chemistry as an integral design space for controlled CRISPR editing. Although RNA chemistry has yet to realize its full potential, it is just as important as enzyme technology in the CRISPR space. Recent publications illustrate significant advances in using modified single guide RNA to turn off or turn on gene editing activity.
For example, a paper in Nature Communications describes how light-sensitive moieties may be used to inactivate editing and thereby temporally control on/off-target events.1 This approach takes advantage of different kinetic rates for on/off-target events and biases editing toward wider safety margins. This approach also enables the spatial control of editing, which may be useful and could be practiced in tissues with current interventional catheters.
Neurodegenerative disease modeling demonstrates the power of iPSCs
Based on previous work in the pluripotent stem cell space, I realized how much I had to learn about developmental biology and disease. Early deterministic events can impact future cell fate or set risk thresholds for later disease. A major difficulty in modeling complex disease lies in recreating or interpreting multigene etiologies and in replicating patient disease cues in a testable preclinical or ex vivo model.
Currently, we have experience in modeling neurodegenerative disease with engineered iPSCs. In these iPSCs, common genetic mutations associated with neurodegenerative diseases are placed in different genetic backgrounds.2 Working with the National Institute on Aging, Synthego is editing iPSCs to introduce mutations relating to Alzheimer’s disease and related dementias. These edited cells will then be used by researchers to better understand these insidious diseases and identify new ways to treat them.
By using iSPCs and studying the effects of placing panels of mutations in different genetic backgrounds, researchers may gain insights leading to new gene-based diagnostics that can detect disease or estimate the likelihood of response to treatment.
Using CRISPR diagnostics and iPSCs to determine treatment response
One research area that is currently taking off and that presents a great investigative opportunity is the use of CRISPR and iPSCs as a model system to integrate rare disease alleles and model treatment and diagnostic approaches. iPSCs are clearly a valuable tool as they can be derived from healthy or patient populations, where genotyping can be used to correlate with function in important dimensions relevant to disease pathology.
Model systems with iPSCs can show how a cell type matures from a primitive germline state. Such systems are useful tools in developmental biology. But model systems with IPSCs can be more elaborate. For example, model systems may consist of iPSC-derived organoids that replicate three-dimensional tissue architectures. These systems may help researchers study how altered genes or viruses contribute to disease, or how patients may respond to treatment regimens.
Increasingly sophisticated iPSC-based model systems align with another area of advancement: the development of artificial intelligence and machine learning technologies to model disease and link clinical health and wellness data back to an iPSC-type rare disease model. This allows for a highly effective iterative testing loop for building rare disease diagnostics and understanding the pathways that are operating in a disease.
Rapid engineering throughput can also enable rare disease modeling by recreating a panel of rare disease alleles in both bulk and single-cell formats. In one case of an inborn error of metabolism, it has been demonstrated that single-cell RNA sequencing can be used to identify the nodes connecting to the rare disease allele. Examination of nodes that change when an allele is reverted or clustered provides a gene-based diagnostic that can stratify patients for personalized treatments or indicate which treatments may obtain the best responses.
Accelerated approval pathways in place to support gene editing
Fortunately, from a regulatory perspective, validated paths for accelerated approval are already in place. The FDA’s Regenerative Medicine Advanced Therapy (RMAT) designation is a regenerative medicine gene therapy–driven initiative that can allow single registrational studies to lead to market approval. This is a sea change in the regulatory environment. It is now possible to guide the development of gene therapeutics with a more direct line of sight to real-world patient access.
Because of the links between gene alterations, analytical measures, and functional outcomes, CRISPR-based gene therapies present opportunities to relate gene correction and durability (tissue genotyping), functionality or potency (gene- or protein-based omics detection), and treatment response (diagnostics based on systems biology mediated from the disease locus). Engineering biology provides the opportunity to build a highly effective translational roadmap for gene therapies, effectively inverting the traditional drug development funnel.
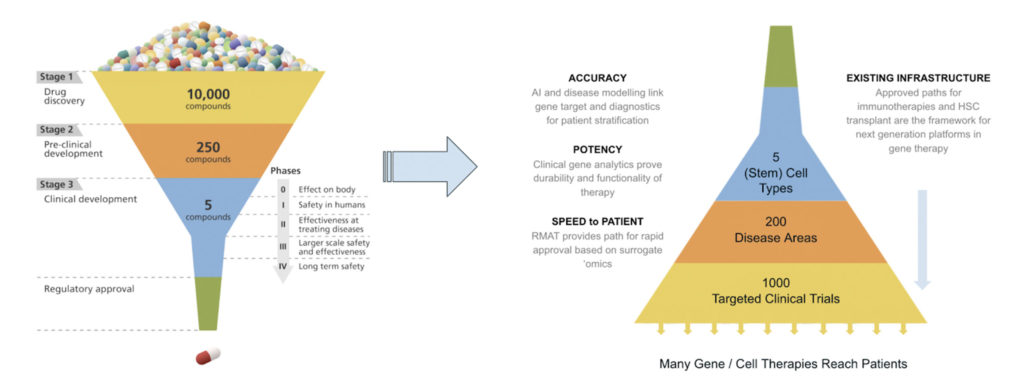
The RMAT approach eliminates, at a minimum, the two parallel Phase III pivotal studies traditionally required for regulatory approval. That in turn reduces by an order of magnitude patient numbers, costs, and the typical 10-year period it takes to conduct those clinical trials.
Besides continuing to represent enormously exciting science, CRISPR will soon realize its promise to reshape our approach to translational medicine. CRISPR-based gene therapies will see exponential growth in the very near term. CRISPR-based genome engineering is becoming a lightning rod attracting a generation of engineers broadening the context of “code.”
References
1. Carlson-Stevermer J, Kelso R, Kadina A, et al.
CRISPRoff enables spatio-temporal control of CRISPR editing. Nat. Commun. 2020; 11: 5041. DOI: 10.1038/s41467-020-18853-3.
2. Synthego Launches High Throughput Induced Pluripotent Stem Cell Genome Engineering. Synthego.com. Published October 22, 2019. Accessed November 24, 2020.
Robert Deans, PhD, is Chief Scientific Officer at Synthego.
To help us preview the future, we asked opinion leaders, all from outstanding technology companies, to discuss a range of new initiatives. The full list of articles is below.
Leroy Hood: Reflections on a Legendary Career
Uncharted Territory: Top Challenges Facing Gene Therapy Development
Envisioning Future Trends in Regenerative Medicine
Engineering Biology—Accelerating Transition
Bioprocessing in a Post-COVID-19 World
Sustainability and the Synthetic Biology Revolution
Sowing the Seeds of Agricultural Biotechnology
Neuroscience Widens Its Investigations of Disease Mechanisms
