When you use CRISPR gene editing on crop plants, you can do “some pretty cool things,” says Timothy Kelliher, PhD, head of crop trait and technology discovery at Syngenta Seeds. You can change the structure of chromosomes, add large amounts of genetic material, move genes around, turn genes on and off, and fine-tune gene expression. And yet, Kelliher admits, inefficiency in bringing these cool things to commercial agricultural products is “still a problem.”
So, why is CRISPR gene editing facing commercial challenges? There are several answers. One of them, Kelliher notes, is the difficulty of modifying plant traits that depend on multiple genes. We’ll hear more about that in a moment. But first, let’s cite a few other complications: plant-specific difficulties in carrying out gain-of-function edits, uncertainties over regulatory distinctions between gene edited plants and GMOs, and modeling limitations that prevent iterative design.
Fortunately, CRISPR crop challenges are being addressed by commercially minded researchers. Several of them have contributed their insights to this article. Let’s start with Kelliher.
Not so simple
Even when important plant traits are known, they may pose daunting gene editing challenges. Often, these traits are complex; that is, they involve multiple genes. “CRISPR is a great tool in a toolbox,” Kelliher points out, “but finding easy ways to edit plant traits with big impacts is rare.”
There are only so many low-hanging fruits (and vegetables) that need only simple modifications to modify a trait. One of the low-hanging fruits is the Arctic Apple, which has, according to popular accounts, been modified to carry a gene that helps reduce browning. Actually, it’s a little more complicated than that. The Artic Apple has been genetically engineered with a transgene that produces specific RNAs that silence the expression of at least four polyphenol oxidase (PPO) genes. (Ultimately, expression of PPO, the enzyme that causes browning, is reduced through RNA interference.)
Browning resistance has also been introduced to a potato, the Innate Potato. Here, as with the browning-resistant apple, the changes that have been introduced are fairly simple—though perhaps not so simple as popular accounts would suggest. For the Innate Potato, fragments of a single potato PPO gene were reintroduced into potato to activate an RNA interference pathway. In addition, a transgene was introduced to use RNA interference to reduce the expression of the asparagine synthetase-1 gene. Limiting asparagine prevents a reaction that occurs in high-temperature cooking and turns asparagine into acrylamide, which has been suspected of being associated with human cancer.
Relatively simple modifications have also sufficed to enhance herbicide tolerance. Often, herbicides kill plants by attacking a specific enzyme, but gene editing can change one amino acid in that enzyme to block the binding of a plant-killing chemical, making a crop resistant to the detrimental effects of the herbicide.
In most cases, though, the optimal alleles for all of the genes involved in a complex trait are unknown. So, even though there are CRISPR tools like Cas12 that allow multiplexing in one construct with multiple guides to different genomic targets, it is a biological challenge to know how to edit each gene involved in a trait and to figure out what effect the different edits have on a trait when they are all put together. What’s more, since there is a great deal of genetic diversity, gene redundancy, and structural variation among the chromosomes in plants, there are, Kelliher says, many examples where a gene or an edit works as desired in one crop line but has a different effect in another crop line.
Less is more

Amfora
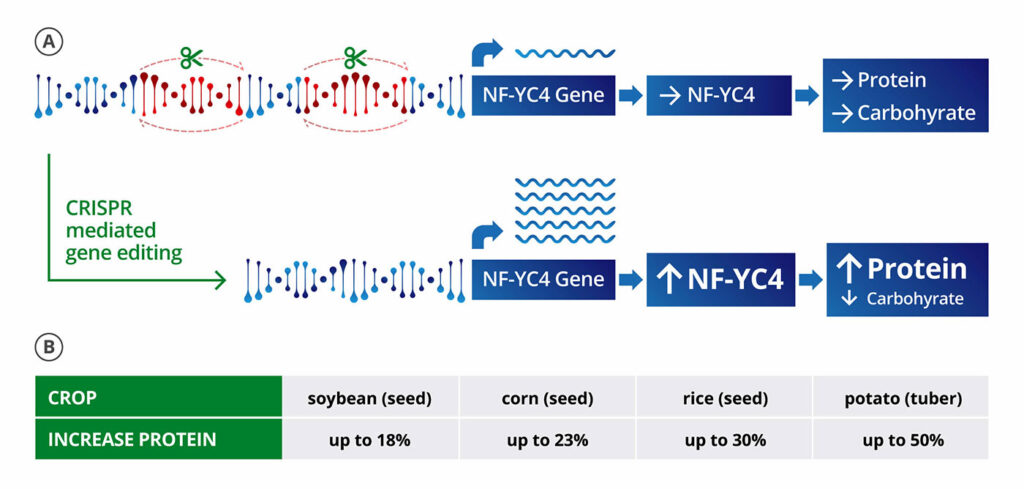
“The simplest application of CRISPR in plants is to make deletions,” says Michael Lassner, PhD, the chief science officer at Amfora, which is using gene editing to improve the nutrition and sustainability of soy crops. “If we can delete negative regulatory binding motifs, it upregulates the gene.”
With the strategy of using deletions to upregulate genes, Amfora is making types of soybeans that have more protein, which is valuable for the main use of soybeans—animal feed. To create a soybean with 10% boosted protein content at the expense of starch production, for example, Amfora is deleting certain regions of the promoter for NF-YC4, a member of a class of transcription regulators that is highly conserved across many plant species, to increase the gene’s expression. (NF-YC4 acts like a switch. When it is on “high,” it favors the production of proteins; when it is on “low,” the production of carbohydrates. Absent specific pieces of DNA, NF-YC4 stays set on low.)
Amfora is working to get to a point where deletions in the promoter regions are passed down through the generations in a stable way, which is critical for developing a new crop. Lassner notes that gene editing has little to no impact on either oil (the other economic product of soybeans) or yield, which is what farmers care about in terms of the cost of goods.
Amfora’s is also developing a soybean product that has “ultra-high” protein content, requires less energy to make, and promises to improve the functionality of soybean meal. “One of the big sources of soy protein is called soy protein concentrate (over 70% protein), Lassner explains. “It gets turned into texturized soy or vegetable protein, which is a key ingredient in a lot of the plant-based meats. And with the crazy commodity prices today, we see there’s a huge amount of room for us to build a business in the food ingredient space by using this ultra-high protein meal.”
Compared to deletions, insertions are much more challenging edits. In animal cells, an allele can easily be swapped at efficiencies of 50–70%, where more often than not the allele gets in correctly. In plants, the efficiency is typically less than 1%. The problem in plants is that the homology-directed repair pathway—the pathway that can incorporate donor DNA—is less readily available. Instead, repair is dominated by the nonhomologous end joining pathway, which simply sticks the loose ends of broken DNA back together.
Overcoming the inefficiency of homology-direct repair in plants is a priority because it would facilitate the introduction of gain-of-function edits, which are desirable because they promise to reduce yield penalties. For example, they could reduce the yield penalty associated with disease-resistance genes in certain rice varieties. When these genes are bred into a new line, they can result in plants that have superior disease resistance but lower yields. So, what if disease resistance could be gained without the yield penalty?
“If we can do that through gene editing, it would be a really powerful new allele that we could deliver,” Lassner tells GEN. “There have been some success stories in that area. That’s next-level stuff that is going to take some technology innovation as well as things like protein structural modeling to create something new that doesn’t exist in nature and then replace the natural gene with this new thing that you’ve designed.”
Introducing new crops
Over the past decade, CoverCress has worked to transform field pennycress, a common winter annual weed, into a novel rotational cash crop through sophisticated breeding and gene editing. “You can use weeds from the roadside and just grow them, but they’re not going to grow as a crop,” says Ratan Chopra, PhD, the company’s vice president of research. “We’ve taken a weed and tried to domesticate it to help farmers grow it as a crop.”

CoverCress
Using CRISPR, CoverCress is focusing on agronomic, compositional, and oil traits that could make pennycress a valuable crop. “The agronomic traits are to get the farmers to believe that it’s a crop and not a weed,” Chopra tells GEN. “In terms of composition, there’s always a meal coproduct with oil seeds. You want the meal for desirable downstream markets, such as animal feed or human foodstuff. Naturally, pennycress has a lot of oil, but to target the renewable diesel industry and the aviation field, we need to improve the profile.”
CoverCress plans a small-scale launch of its gene edited pennycress product this year. But the company doesn’t expect to bring a human foodstuff to market any time soon. “We are staying away from food markets until regulatory agencies figure out what needs to be done with the new crop and how they’re going to handle it,” Chopra relates.
That’s not the only regulatory hurdle that CoverCress is trying to navigate. Some regulatory bodies have put up restrictions for the way gene edited crops are developed. For example, two genes cannot be targeted simultaneously in the United States without a Regulatory Status Review, which can take anywhere between one and two years, and which limits the use of multiplexing CRISPR technologies like Cas12 that can target multiple genomic loci with one construct.
“You cannot use two guides with the editing technology to target two genes simultaneously,” Chopra explains. “In one construct, you can do only one gene at a time and then cross them to each other. So, that’s a bottleneck. It’s not because of the technology; it’s about the way the regulators are thinking about products and how you create them.”

GMOs vs. gene edited plants
All of this CRISPR-driven innovation in agricultural biology hinges on the regulatory decisions made by the governing bodies around the world. And everyone is holding their breath as to whether these gene edited plants will be classified as genetically modified organisms (GMOs).
Recently, on February 7, 2023, Europe’s highest court said that the European Union’s laws restricting the use of GMOs would exclude in vitro plant gene editing techniques, provided they are used conventionally and have a long safety record. The decision appears to be part of a trend observed by Dan Jenkins, head of regulatory and government affairs and quality at Pairwise.
“Many governments have and are preparing to come to the decision that a lot of the applications of this technology and plants are not considered GMOs,” he says. “The governments in South America, Central America, Japan, and India are saying these applications aren’t like GMOs. I would say that the way regulations are right now around the world is very positive and encouraging.”
There are also regulations to make sure that the gene edited products are safe, and Pairwise indicates that it has incorporated a lot of regulatory and food safety elements to develop a fresh, high-quality, safe, nutritious product for consumers. However, Jenkins believes that what should matter is the end product, not how it was attained. Jenkins explains that people have been successfully selecting traits in plants through breeding over hundreds and thousands of years.
For example, mustard greens, cauliflower, Brussels sprouts, and kohlrabi all come from selecting for traits in the Brassica genus of vegetables. “Those are pretty different-looking things, and the history of safety with those pretty substantial changes shouldn’t give anybody too much cause for worry, rather the opposite,” Jenkins maintains. “Ruby Red Grapefruit was created through mutagenesis, and that’s a pretty healthy thing to eat. I think it should be about the product, not the process.”
Improving in silico models
Some companies have built in silico models that use machine learning to predict allelic combinations that would, if implemented, have a high likelihood of providing a beneficial outcome toward a desired trait. These companies test plants that possess these combinations, and then the results are passed back to the models. According to Kelliher, most of the currently available models identify genetic markers on chromosomes that are important for traits such as yield.
The models do not actually recognize which genes are involved. For example, the models might say that if you ensure a beneficial genomic marker at one chromosomal location is accompanied by a similar marker at another chromosomal location, you will increase your chances of getting an even better yield. Models that use only genomic markers satisfy breeders, who do not need to know which genes are actually involved. However, such models are inadequate for developers who are interested in genome editing. These developers need to know which genes and pathways are implicated.
Kelliher says what the agricultural biotechnology field needs to help guide CRISPR use is a discovery platform that’s efficient and tests things in silico with models that recognize genes, metabolic processes, or signaling pathways, and that get retrained and strengthened by in vivo testing to the point where they can actually predict the two or three genomic changes that will provide the trait outcome. Such models could help companies have a massive impact on developing and breeding the produce of tomorrow.

