Cancer is caused by changes in DNA that lead to uncontrolled cell growth. Some of these changes correspond to inherited or germline mutations; most correspond to acquired or sporadic mutations. Cancer-causing DNA alterations include single nucleotide variants (SNVs), copy number variants (CNVs), insertions/deletions (INDELs), and rearrangements.
When these DNA alterations are identified, they can be exploited as biomarkers and drug targets. They can also be used to realize the promise of precision medicine. That is, the DNA alterations unique to a given patient’s genome could indicate which targeted treatments would be effective.
Unfortunately, our knowledge of cancer-causing DNA alterations is far from comprehensive. But it is improving, thanks to projects such as The Cancer Genome Atlas (TCGA). The project, a joint effort of the National Cancer Institute and the National Human Genome Research Institute, was completed in 2018. It analyzed 20,000 tumors representing 33 types of cancer, and it generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. All of this data is now publicly available to the research community.
Many current efforts reflect an interest in using genomic techniques to advance diagnostic testing and translational research applications. Outstanding examples of these efforts were showcased at the Cancer Genomics Consortium’s 12th Annual Meeting last August. Among the presenters were experts in sequencing, genomics-based tumor detection, and other technologies that are advancing the discovery of pathogenic variants, the screening and stratification of patients, and the development of new drugs.
An integrated NGS workflow for lung cancer
At the meeting, a case study of a lung cancer sample was presented by Brigette Brown-Kipphut, a field application scientist manager at Agilent Technologies. She noted that the case study relied on a next-generation sequencing (NGS) workflow that Agilent had developed, a workflow that accommodates the extraction of nucleic acids from tumor biopsies.
Brown-Kipphut reported that immunohistochemistry analysis revealed that the sample had PD-L1 expression in at least 50% of tumor cells, and that fluorescence in situ hybridization was negative for an ALK/EML4 rearrangement and ROS1 fusion. Sanger sequencing was negative for epidermal growth factor receptor hotspot variants. The cancer was identified as a nonsquamous non-small cell lung carcinoma (NSCLC).
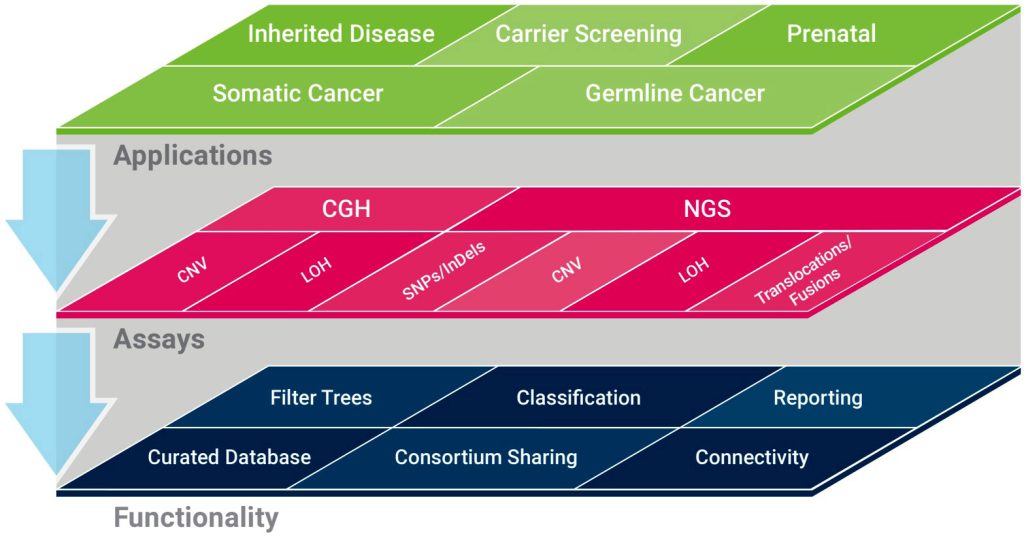
At this point, one could test for further mutations such as rearrangements and CNVs, Brown-Kipphut stated. But doing so could be time consuming and expensive, with no guarantee of turning up a useful result. So, Brown-Kipphut suggested an alternative: one could use NGS to streamline the process and generate highly comprehensive information about the tumor genome.
To take this alternative approach, one could use Agilent’s SureSelect Cancer All-in-One Lung Assay, a targeted NGS assay that can analyze a lung cancer panel of 20 genes relevant to NSCLC. Brown-Kipphut emphasized that the assay consolidates the detection of all variant types into one workflow. When sequencing is complete, variants are called and
analyzed in the context of the scientific literature and public cancer genome databases. Finally, a report is generated.
In the analysis described by Brown-Kipphut, the sample was found to have an ALK/MYO16 rearrangement between chromosomes 2 and 13 and an ERBB2 amplification. These results, she argued, demonstrate that NGS technology can significantly reduce costs and turnaround times from sample submission to result.
Depending on the number of tests that NGS replaces, an analysis that usually takes one to four weeks can be performed in three to five days, Brown-Kipphut asserted. “Since lung cancer tumors are often heterogeneous, variants may be low in frequency,” she added. “The use of the molecular barcodes in this assay allows for the detection of those variants previously too low to identify because of background noise or PCR/sequencing errors.”
Deeper genomic analysis
Emily Paul, PhD, product director for SOPHiA Genetics, presented a “deep dive” on solid tumor detection. She outlined how the company’s DDM platform expands the scope of NGS-based testing beyond the detection of variants such as SNVs and INDELs. DDM also assesses immuno-oncology biomarkers such as microsatellite instability (MSI), the molecular fingerprint of a deficient mismatch repair system.
SOPHiA asserts that one of its DMS-based solutions, the Solid Tumor Solution (STS), can achieve 100% analytical sensitivity and specificity when it is benchmarked to the PCR gold standard. The solution’s algorithms have been expanded to accommodate results from comprehensive testing, to take in more NGS data points for assessment of stability, and to evaluate a variety of tumor types.
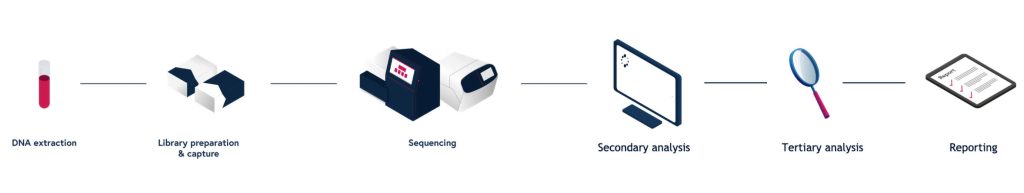
and streamlined end-to-end workflows.
SOPHiA’s DDM platform also facilitates the determination of tumor mutational burden (TMB) scores, the total number of mutations per coding area of the tumor genome, in comprehensive genomic assays. TMB is important because it can be an indicator on how the body’s immune system will respond to the tumor itself as well as the growing number of treatments in the immuno-oncology space.
“The small multigene panel tests are crucial,” Paul insisted. “They are important because they help us understand hotspot mutations. But we’re growing in our understanding and what we want to query. If we just did small panels, we would miss out on hundreds of other targets that can indicate the success or efficacy of a treatment.”
SOPHiA is part of a trend toward comprehensive genomic profiling. The company, Paul noted, has developed technology that can do more than test for just a handful of genes. It can test for over 500 genes simultaneously in a comprehensive genomic profiling assay.
Paul added that on the clinical side, SOPHiA is evolving into the precision medicine space. “We have an ongoing clinical study to perform a retrospective analysis on the confluence of imaging data, genomic data, and treatment data for stage IV NSCLC,” she elaborated. “By applying our machine learning algorithms to these data sets, we can stratify patients based on progression-free survival. This can inform the treatment decision, scaling up or down the intensity required for effectiveness.”
Personalizing cancer genomics analysis
Circulating tumor DNA (ctDNA) is another way of probing the cancer genome. It can be used to monitor cancer status during therapy or to detect cancer recurrence after therapy. These possibilities were discussed at the Cancer Genomics Consortium meeting by two presenters from Natera, a specialist in ctDNA testing.
John Simmons, PhD, global vice president for biopharma business development, and Angel Rodriguez, PhD, medical director for oncology, described Natera’s platform, Signatera, and how it enables a personalized, tumor-informed ctDNA assay for detection of molecular residual disease assessment and recurrence monitoring. Simmons said the assay starts with the extraction, whole-exome sequencing, analysis, and filtering of genomic DNA from tumor tissue and matched normal whole blood. This procedure identifies patient-specific somatic mutations. Essentially, it produces what Natera calls a tumor barcode.

Next, an algorithm selects tumor mutations to follow in the patient’s blood that can be used in a multiplex PCR assay. “The top 16 somatic variants are selected based on clonality, detectability, and frequency of mutations,” according to Natera. “Multiple-PCR compatible primers are designed for each of the 16 somatic variants.” Ultimately, the patient’s blood samples are processed for multiplex PCR amplification and ultradeep sequencing to detect the presence of ctDNA.
“In the larger majority of solid tumors, the level of molecular fragments of DNA in the blood is extremely low,” Rodriquez noted. “With previous technologies, it really has been impossible to detect. Now, with our approach, where we are creating this molecular barcode of multiple mutations unique to each patient’s cancer, it allows us to detect disease at much lower levels than we ever have in the past.”
Natera validated its test against a collection of blood samples from patients with many different cancer types, including colon, breast, lung, bladder, esophageal, and ovarian cancer, and the company determined that Signatera was able to detect disease recurrence with 90–95% accuracy. “The take-home message,” Rodriguez emphasized, “is that we can find evidence that the disease is going to recur an average of eight months before it actually starts to cause symptoms.”
Natera is testing Signatera in separate clinical trials in partnerships with
Genentech and other pharmaceutical companies for the detection of residual disease in patients who have completed cancer therapy.
Writing the future of cancer research
Twist Bioscience has taken cancer genome research in yet another direction by targeting malfunctioning proteins with custom therapeutic antibodies designed from scratch with its DNA writing technology. The company’s CEO, Emily Leproust, PhD, said that the core of the technology is a silicon chip the same size as a standard 96-well plate. But instead of having 96 wells, the chip has one million DNA sequences printed on it like rows of tiny Lego towers. It is, essentially, a silicon-based DNA synthesis platform.
Twist uses its platform to make products of different types. When making antibodies, for example, Twist uses the platform to synthesize antibody-coding DNA sequences (more specifically, sequences corresponding to antigen-binding regions). These sequences are incorporated into a gene-to-protein workflow that produces antibody libraries.
The libraries make it possible for Twist to screen 10 billion different antibodies against a target of interest. The company takes the best hits from the first round of screening and introduces anywhere from 1 to 10 mutations to create a second round of one million DNA sequences. “Typically, we end up with an antibody that is extremely powerful,” Leproust asserted. “It’s an antibody that binds really well and has great function.”
The result is a novel method of drug discovery. Selling pharmaceutical companies on the idea, though, turned out to be challenging because it sounded too good to be true. Leproust recalled what Twist started doing to get a foot in the door. Whenever Twist approached a company, it would propose a project that would solve the company’s most challenging problem. For the company, such a project would represent a low-risk proposition.

“So far, we have a 100% success rate,” Leproust declared. “They give us the hardest of the hard, and [our approach] works when they have failed. After that, they give us the easy stuff and they’re willing to pay more.” Twist has now racked up dozens of successes in delivering high-affinity, high-functionality, fully human therapeutic antibody candidates to pharmaceutical industry partners. Cancer targets Twist has worked with include A2A, CD3, PD1, TIGIT, and CXCR4—plus two other undisclosed cancer targets that Leproust indicated are available for licensing.
Expediting variant detection
Detection of somatic variants through hybridization capture typically requires one and a half days of total turnaround time due to the performance gain of an overnight hybridization reaction. To support a single-day workflow, Roche has developed the KAPA HyperPETE product portfolio of reagents and kits. KAPA
HyperPETE products can be used to detect all major somatic variant classes, including SNVs, CNVs, short INDELs, MSI status, and fusion transcripts (including unknown fusion partners). The single-day workflow can combine the superior performance of hybrid capture with the speed and simplicity that has been possible only with amplicon workflows.
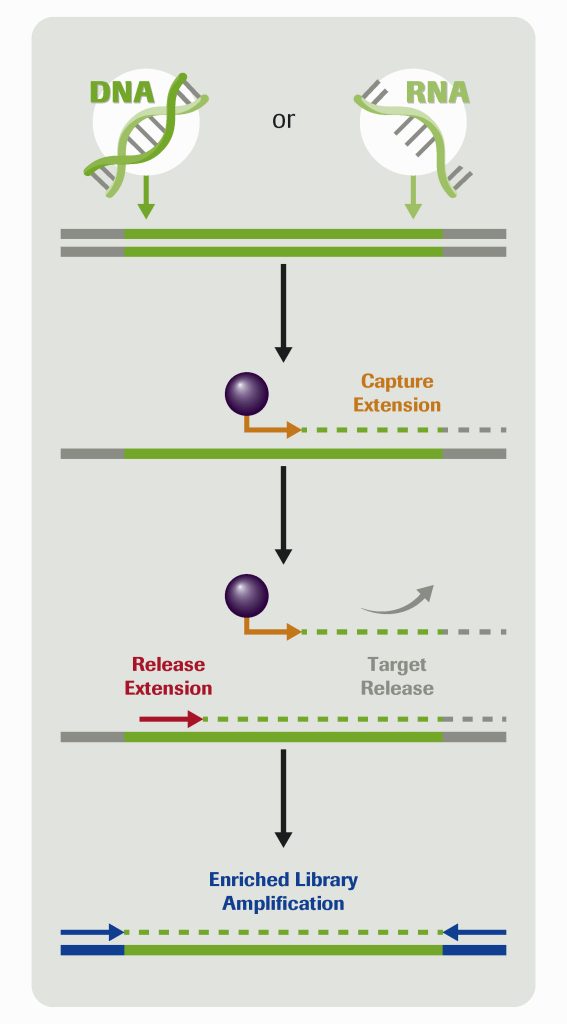
As a way of illustrating the utility of the workflow, Brian Godwin, director of reagent development at Roche, presented preliminary data from KAPA HyperPETE somatic oncology panels. For example, he indicated that the KAPA HyperPETE Pan Cancer Panel was used in a somatic cfDNA workflow. The assay detected 100% of short variants at 1% allele frequency across all reference cell line DNA samples.
A key to the workflow is making use of a target-specific biotinylated primer extension with DNA polymerase, which Godwin said quickly drives reactions forward. That step takes 30 minutes. The resulting primer extension complexes are captured to streptavidin beads while off-target library molecules are washed away. Then a second target-specific primer extension step releases molecules off the beads where they can be collected and sequenced.
“We get specificity out of the capture primer extension,” Godwin asserted. “But we also know that adding the second release primer extension is going to give you added specificity, so you sequence the targets that you’re really interested in.”