The term “gene” was introduced in 1909 to refer to the fundamental unit of inheritance. Then, just a few years later, in 1914, the existence of cancer-causing genes was predicted to account for the origin of tumors. Eventually, in the 1970s, researchers began identifying individual cancer-causing genes, including mutated proto-oncogenes and mutated tumor suppressor genes. By now, many such genes are known.
Although the recent genetic discoveries are encouraging, it is apparent that many cancer-causing genes have yet to be discovered. Also, it has become clear that cancer cases typically involve multiple cancer-causing genes. Cancer, then, is more complicated than a one-to-one relationship between mutation and tumor. Indeed, different genes have been implicated in the origin of cancer in different cells within the same tumor.
If we are to understand cancer, we must sustain a comprehensive approach to cancer genetics. One way we can do so is to extend the kind of work that was pioneered in the Human Genome Project. For example, we can derive insights from The Cancer Genome Atlas (TCGA), a massive cancer-fighting effort that was supervised by the National Human Genome Research Institute and the National Cancer Institute. The project established a publicly available catalogue of cancer-causing genomic alterations.
TCGA data sets and other resources are helping to improve cancer diagnosis, treatment, and prevention. Nonetheless, many challenges in cancer genomics remain, and they will not be overcome unless the most advanced tools and technologies for cancer genome profiling are put into use. Such tools and technologies were featured at the Cancer Genomics Consortium’s 12th Annual Meeting, a virtual event that was held last August.
Some of the meeting’s outstanding presentations are revisited in this article. For example, this article describes next-generation sequencing (NGS) tools that integrate with automated workflows, require minimal amounts of starting material, and generate comprehensive genomic profiles in a single day. This article also relates that automation is being used to expedite critical cytogenetic procedures such as cell harvesting, slide preparation, and staining. Yet another feat of automation reported in this article is the miniaturization of processes such as fluorescence in situ hybridization (FISH).
Finally, this article highlights a noninvasive approach to testing for colorectal cancers. This approach relies on a unique methodology, specifically, a multitarget stool mRNA test.
Smaller samples, more NGS information
Clinical analysis of patient samples is often hampered due to the amount of material needed for complex multianalyte investigations. Luca Quagliata, PhD, BCMAS, global head of medical affairs, clinical next-generation sequencing, and oncology at Thermo Fisher Scientific, argued that any tool that can work with limited input can help researchers discover more information from available specimens. That’s one reason why the company is focusing on developing NGS assays and solutions that can deliver comprehensive genomic profiling from minimal starting material.
“Lower input requirements mean more samples and smaller amounts that can be tested to accelerate biomarker discovery and guide the development of future treatments,” Quagliata explained. “We believe that no samples should be left behind, ensuring no patient who is contributing to advancing science will be left behind.”
The company’s latest NGS technology, the Ion Torrent Genexus System, is the “only fully automated NGS-based platform that can provide results in a single day,” Quagliata asserted. “With automated NGS, researchers can spend less time preparing and processing samples so they can focus on analyzing the data and advancing scientific research.”
Traditionally for NGS, a nucleic acid sequence is fragmented into smaller parts that are subsequently amplified and bound to beads. With the Ion Torrent NGS approach, the beads are placed into millions of tiny wells on a semiconductor chip—arrayed like the light-sensitive parts of a smartphone camera. However, instead of detecting pixels of light, the Ion Torrent Genexus System detects hydrogen ions. As hydrogen ions are released from each well, the pH change is simultaneously measured to determine the sequence of each fragment. “The chip,” Quagliata remarked, “essentially contains millions of pH meters that tell us what base pairs are present in each fragment.”
One of Thermo Fisher’s accompanying new technologies is the NGS Oncomine TCR Beta-LR Assay. It takes a multidimensional approach to profiling tumor immune interactions.
Quagliata elaborated, “Studies have found that T-cell receptor repertoire analysis outperformed established biomarkers (such as PD-L1) as a predictor of response to chemo/immunotherapy in lung cancer. Deciphering the role of the immune repertoire is an exciting new area of cancer research.”
Overall, Quagliata believes that new strides in NGS technologies will concomitantly improve applications across oncology, infectious disease and microbial research, reproductive health, human identification and forensics, and agriculture. He reflected, “Oncology offers one of the largest areas of opportunity as the unique complexity of the cancer genome is increasingly linked to treatment response and outcomes.”
Cytogenetics automation
Diseases such as cancer often feature chromosomal breaks, rearrangements, and added or missing segments. Cytogenetic (that is, chromosomal) analyses can help clinicians improve their diagnostic capabilities, treatment decisions, and outcome assessments.
“Certain processes in the cytogenetics workflow, which for many years have been performed manually, are slow and labor intensive,” said John McCloskey, director of sales, North America, ADS Biotec. “There is an ongoing shift by many laboratories to adopt automation for workflow tasks that occur upstream of analysis and pathology review.
“This shift to automation is born from the need to maximize the productivity of highly skilled technicians and technologists given the diminishing ability to attract and maintain such personnel. This challenge has been further exacerbated by the COVID-19 pandemic.”
ADS Biotec develops, sells, and supports automated systems and consumable kits for cytogenetic, pathology, and clinical R&D laboratories. Process steps involved in cytogenetics workflows include cell harvesting, slide preparation, and slide aging/staining. The company has developed automated solutions for each of these processes.
For example, ADS Biotec’s HANABI Metaphase Chromosome Harvester system can automate the harvesting process from centrifugation through final fix. Downstream of harvesting, dropping and spreading operations are performed within an environmentally controlled chamber. These operations can be automated using the company’s HANABI Auto-Spreader and BioDot instruments. Subsequently, slides can be aged and stained using the HANABI UV Aging device and the HANABI Stainer.
McCloskey advised that laboratories should consider performing an audit to estimate the costs and benefits of investing in automation: “By automating key processes in the cytogenetics workflow and enabling the reallocation of personnel, [laboratories can measurably improve] workflow efficiencies, turnaround time, and output. Further, it is important to consider consistency, reliability, and enhanced safety that automated technologies provide.”
FISH-ing for ultralow volumes
Challenges for today’s cytogenetic laboratories include the need to lower reagent costs, standardize processes, and reduce hands-on times. To help laboratories overcome these challenges, instrument providers are offering more efficient technologies. BioDot, a global provider of high-throughput, ultra-low-volume dispensing systems, has introduced the CellWriter S2 to automate slide preparation and slide processing for FISH and chromosome analysis.

“The CellWriter S2 achieves unparalleled reductions in reagent costs through miniaturization of FISH in a nanoliter reaction utilizing just 0.35 microliters of diluted DNA probe per reaction,” reported Matthew Sergent, BioDot’s director of sales, molecular diagnostics. “When compared to manual processes using 3, 5, or even 10 microliters of probe, this provides unprecedented savings for laboratories.”
Sergent said that although most laboratories require costs savings, even more important drivers for exploring automation are time savings and standardization. “Cytogenetic laboratories continually report growth in assay volumes outpacing staffing capacities in a highly competitive labor market,” he explained. “Unlike any system on the market, the CellWriter S2 meets the needs of cytogenetic laboratories in a single instrument that fully automates slide preparation and processing.”
According to Sergent, cytogenetic laboratories have long struggled with intermittent quality issues due to personnel changes or seasonal changes. (The latter are generally deemed more important.) He advised, “The CellWriter S2 has been shown to significantly reduce the variability that is often seen over a period of time or between technologists—regardless of experience—by enabling a consistent slide preparation process that repeatedly provides high-quality results.”
After having struggled to meet rampant demands for COVID-19 testing services, many laboratories are finding that they have exhausted their capital budgets for new equipment. Sergent said that BioDot is addressing this by introducing an industry-first use model with no capital expense. He declared, “We see this as a unique opportunity that allows laboratories to take advantage of the most advanced automation on the market while simultaneously achieving unprecedented savings through reagent miniaturization.”
Optical genome mapping
Cancer research involves multiple technologies used either simultaneously or in a tiered fashion to impart a comprehensive analysis of the neoplastic genome. However, about 50% of malignancies are cytogenetically normal, according to Alka Chaubey, PhD, FACMG, chief medical officer, Bionano Genomics. She advised, “Optical genome mapping (OGM) technology provides an alternative that can better stratify these malignancies for clinical research as well as improve the success rate of finding structural variants (SVs) for better cancer management.”
The company’s Saphyr OGM system can reveal large SVs that are missed by NGS. “OGM detects SVs ranging from 500-base-pair to megabase-pair lengths and offers assembly and discovery algorithms that far outperform sequencing-based technologies in sensitivity,” Chaubey detailed. “For mosaic samples or heterogeneous cancer samples, OGM detects all types of SVs down to 1% variant allele fraction. Using ultrahigh-molecular-weight DNA, OGM can also detect complex rearrangements and repeat regions of the genome that contribute to various genetic diseases.”
Saphyr works by imaging ultralong, linearized DNA molecules labeled at specific six-base-pair sequence motifs. Comparative analysis of the label patterns over long contiguous reads across the whole genome reveals SVs (>500 base pairs) at sensitivities as high as 99%, with false-positive rates below 2%. Chaubey summarized, “All major types of large structural variants can be detected, even at allele fractions as low as 1%, which is not enabled by any other genomics technology.”
Noninvasive multitarget RNA testing
Although DNA mutation analysis can predict hereditary risk or signal disease development, it cannot provide crucial information on phenotypic or quantitative changes. By contrast, real-time RNA analysis can yield better assessments of DNA variants and health status. Geneoscopy is focusing on noninvasive multitarget RNA testing for colorectal cancer (CRC), the third leading cause of death in the United States.

Chief Science Officer, Geneoscopy
“Nearly 40% of patients remain unscreened for CRC, [presumably because of] the inconvenience and invasiveness of colonoscopies,” noted Erica Barnell, PhD, the company’s chief science officer. “In addition, COVID-19-related cancellations, delays, and backlogs of nonurgent medical and surgical procedures, including colonoscopies, continue to impact CRC prevention and early detection efforts.”
Barnell, who is pursuing a medical degree at the Washington University School of Medicine, added, “Geneoscopy’s RNA-based noninvasive multitarget platform uses stool-derived eukaryotic RNA (seRNA) biomarkers that, when combined with other stool-based biomarkers, have demonstrated better sensitivity in detecting colorectal neoplasia relative to existing commercial FIT (fecal immunochemical test) or FOBT (fecal occult blood test) diagnostics.”
RNA from stool samples can be degraded or contaminated by bacterial RNA, making it unviable for downstream high-throughput analysis. Through Barnell’s research, the company has developed a proprietary extraction method that eliminates bacterial noise, enriches host signals, and effectively preserves intact host RNA.
In 2020, the company’s preventive screening technology earned the FDA’s breakthrough device designation. In a press announcement, the company credited the technology’s ability to detect advanced adenomas with high sensitivity: “[Early] detection of advanced adenomas [facilitates] the removal of precancerous lesions that have the highest propensity for malignant transformation.”
Geneoscopy is conducting a 10,000-person clinical trial called CRC-PREVENT. “This trial will evaluate individuals over the age of 45 who are at average risk for developing colorectal cancer,” Barnell detailed. “Data from this clinical trial will support a premarket approval submission to the FDA.”
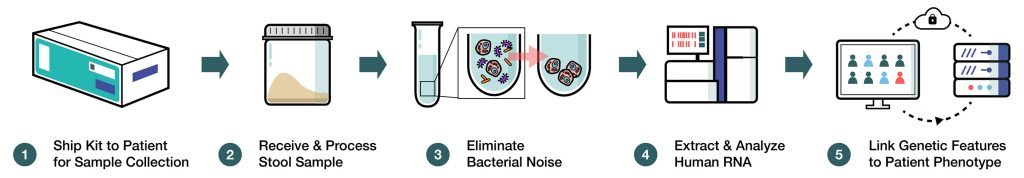
Barnell added that the company’s RNA extraction methodology also could be utilized for screening, diagnosis, therapeutic selection, monitoring, surveillance, and drug development.
“The technology is potentially useful for a host of other diseases including inflammatory bowel disease, infectious disease, necrotizing enterocolitis, celiac disease, irritable bowel syndrome, and diabetes,” she concluded. “Our ultimate goal is to improve the prevention, detection, and treatment of gastrointestinal disease across this entire continuum.”

