The genome is the informational core of the life sciences. That’s why the genome is a source of wonder—and frustration. Yes, the genome is densely packed with valuable information. But it must be unpacked. And if unpacking the genome seems a painstaking business, consider that when we say “genome,” we actually mean “genomes.” That is, we often find it necessary to sift through many genomes to identify meaningful similarities and differences.
Genomes would be hard to analyze even if they were sequenced and stored with any consistency. But they’re not. Genome sequencing and storage are notoriously variable. So, how might genome analytics find solid footing on ground that seems so fluid? With artificial intelligence (AI).
AI technologies such as machine learning (ML) and deep learning are already appreciated for their prodigious data processing and pattern recognition abilities. These technologies, however, are beginning to demonstrate additional abilities. For example, they are getting better at coping with idiosyncrasies.
In genome science, there are a number of obvious idiosyncrasies. These include differences in sequencing protocols and technologies, as well as in data storage formats and data sharing practices. Less obvious idiosyncrasies, however, may be just as important. These include all the complications that may arise when genome analysis attempts to relate genome data to other sorts of data, such as phenotypic data, or data from other omics disciplines.
The idiosyncrasies of genome science are seen as opportunities by AI specialists. Several of these specialists are discussed in this article. Some develop proprietary AI systems for in-house drug discovery or diagnostics applications. Some develop systems to facilitate the sharing and mining of data by a wide variety of organizations. Both kinds of systems have roles to play in uncovering disease mechanisms, developing novel drugs, and improving healthcare.
Democratizing sequencing
Traditionally, an expert’s touch was needed to fine-tune the algorithms that derived meaning from sequencing data. That was all well and good so long as data sets were reasonably small and consistent. But what happens when data sets assume gargantuan proportions and incorporate data of varying quality?
“To make sure you have the highest accuracy and sensitivity, an AI/ML-based system that trains on data sets is better at detecting nuances and figuring out how to make a variant call than humans,” says John Ellithorpe, PhD, president, DNAnexus. “AI democratizes the sequencing field. You do not have to understand exactly what the error profile from the sequencers looks like. Instead, you may expose enough to the algorithm to have a good distribution of the data, and then the algorithm figures it out.
“A lot of data is crucial to get high-quality downstream results. If the use case is not present in the data set, it is impossible to train a machine to find it.”
DNAnexus offers two main products: Titan and Apollo. Titan focuses on high-throughput genomic data processing to identify variants and mutations and to build up large, high-quality genomic data sets. (Titan is used heavily in the diagnostic space, but it is also used to support discovery.) Apollo enables users to derive insight by performing complex studies on large genomic data sets, combining genomic and phenotypic data from clinical trials, medical records, and other sources, to identify and correlate mutations with a specific disease outcome, disease progression, or other factors to aid researchers looking for new drug targets.
The use of AI for association work is still at an early stage according to Ellithorpe. ML is well suited to finding patterns and relationships in data, it but needs sizable data sets to reduce bias and increase sensitivity. The aspiration is that once large high-quality data sets are available, ML algorithms will better identify correlations and determine causal relationships.
“Healthcare data need to be in a secure and compliant space before processing,” Ellithorpe adds. “We help customers bring the data into this space where they process it with a single platform.”
DNAnexus was awarded a research and development contract by the U.S. Food and Drug Administration (FDA). The agency’s Office of Health Informatics wants DNAnexus to build precisionFDA, an open source community platform for using genome analysis to advance precision medicine.
Enabling collective understandings
Too often, biomedical data is inaccessible or unusable. It may be squirreled away, or it may be scattered about. Making all this data available and useful is the self-appointed task of Lifebit, a company that has developed a federated genomics platform that incorporates an AI-enabled real-world evidence (RWE) analysis system.
According to Lifebit, the company’s platform is federated in the sense that it makes all types of biomedical, omics, and clinical data—and RWE—easily accessible and usable. Regardless of where data sets reside, the platform sits on top of them, standardizes them, and makes them appear as though they were all in one place. RWE, it should be noted, includes data harvested from scientific publications and other open sources.
Lifebit emphasizes that its mission is to empower biomedical data custodians to make their data findable and usable for data consumers. Lifebit also gathers RWE from across the web, social media, publications, blog posts, and more, and then combines it with genomics and clinical data. As data sources are updated daily, the system constantly learns. Lifebit also transforms unstructured clinical data, such as images, into structured usable data.
“A majority of our clients make their data available to all other authorized users,” says Maria Chatzou Dunford, PhD, co-founder and CEO, Lifebit. “Different types of organizations that accumulate large volumes of data that we call data custodians have a big interest in making these data available for clinical research and drug discovery.
“Many of our clients are interested in which diseases are emerging (and where) to prevent or be prepared for the next epidemic and also to understand which diseases are most prevalent. They want to see if existing drugs are available for the indications, or if they need to develop new therapeutic approaches.”
Although the largest portion of the platform focuses on human health, it is also being used to surveil animal health. For example, Lifebit technology is helping Boehringer Ingelheim track data related to animal diseases.
Removing bottlenecks
“One of the reasons the cost of drug development has grown so much despite massive efficiency improvements is the reliance on reductionist models, especially for very complex diseases,” says Alice Zhang, co-founder and CEO, Verge Genomics. “The key to transforming the cost of drug development is not expanding the funnel but removing the bottleneck by predicting human effectiveness earlier in the process.”
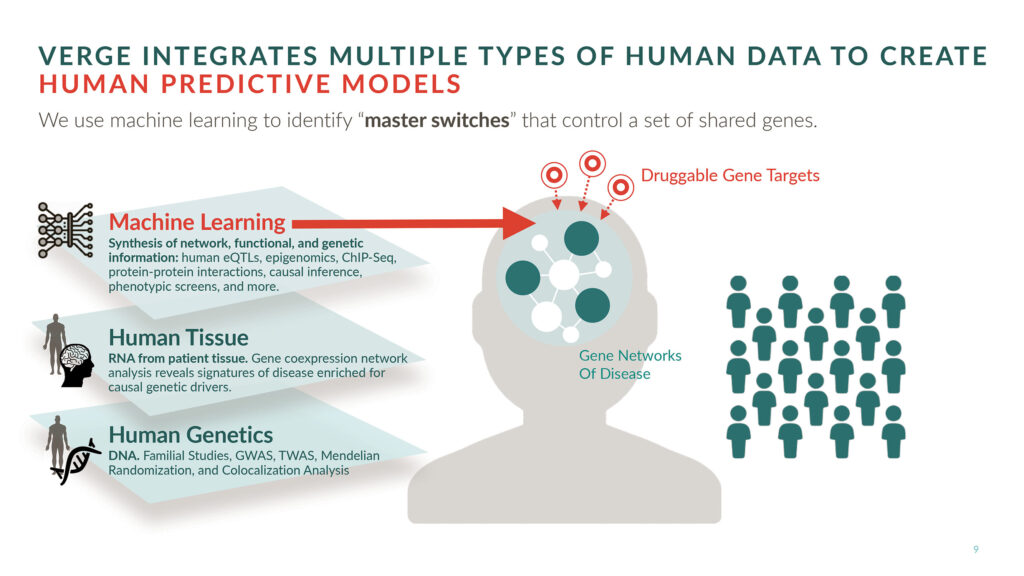
A full-stack, AI-driven therapeutics company, Verge integrates human genomics with ML to develop drug candidates for complex neurological diseases such as amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Parkinson’s disease. The company’s lead ALS candidate will enter the clinic next year.
Verge advocates an “all in human” approach to drug development. That is, the company believes that if one wants to succeed in humans, one should start in humans. At the heart of Verge’s development platform is a proprietary human genomics database that was built over the last three years by partnering with dozens of brain banks, hospitals, and universities worldwide.
“We have collected thousands of different brain tissues directly from patients and RNA sequenced them internally to measure the expression of all of the genes in their genomes,” Zhang details. “We then mine these data with our internally developed ML algorithm to predict therapeutic targets.”
To understand how genes relate to disease, Verge studies disease signatures, which consist of expression profiles that encompass hundreds of genes. Rather than focus on single genes in isolation, Verge considers how single genes may act as master switches that effectively turn disease signatures on or off.
Once master switches in the human genome are identified, they are validated as targets in human-derived neurons. These are neurons that Verge generates from its large repository of human stem cells. Stem cells for the study of a particular disease come from patients who have that disease.
“Neurodegenerative diseases are the biggest medical challenge of our generation,” Zhang declares. “Historically, people have treated neurodegenerative disease as a single disease and looked at common clinical end points. What we are recognizing is that there are many patient subpopulations with different genetic causes.”
The state of knowledge for neurodegenerative disease is where that for cancer was 10 years ago, and it is essentially ripe for the same transformation.
According to Zhang, the biggest revolutions in a field occur when that field applies technologies from other fields. Zhang believes that if the field of drug development applies AI, it will overcome persistent difficulties such as biological complexity and the lack of adequate preclinical models.
Accelerating discovery
An RNA therapy essentially consists of an RNA molecule that is somehow shielded from degradation. Protective options include chemical modifications and delivery vehicles such as nanoparticles and viral vectors. These allow the RNA molecule, a sequence of nucleotide bases, to reach the cell and exert its effects.
RNA therapeutics include small interfering RNA (siRNA), which can completely silence a gene’s expression, and microRNA (miRNA), which reduce a gene’s expression. In either case, the therapeutic RNA’s effects depend on the targeting of a particular gene’s corresponding messenger RNA (mRNA), with the targeting determined by the RNA therapeutic’s base sequence. Because base sequences amount to little pieces of code, they lend themselves to computer analysis.
“Manually sifting through data does not produce the highest quality targets and compounds, nor does it scale,” says Brendan Frey, PhD, founder and CEO, Deep Genomics. “Winning the race to develop RNA therapies requires mastering the enormous complexity of RNA biology, and the only way to do that is with AI. Computers are much better than humans at quantitatively prioritizing targets.”

“Data alone do not produce drugs,” Frey insists. “It was thought that once the whole genome was sequenced, drugs would just fall out. That did not happen. Going from data to a drug requires inference making based on logic, pattern matching, and reducing and expanding data—and more. Inference making is the hard part.”
To make inferences, the human or AI thinking machine must be able to look at the data in a way that is useful. “Transforming publicly available data into a new data set that AI can use is a very difficult science and engineering task that Deep Genomics excels at,” Frey asserts.
Deep Genomics’ AI workbench includes built-in predictors that indicate which genetic variations will affect different processes in RNA biology, and how a potential therapeutic compound will fix the problem.
Over 40 ML predictors enable the company to address varied drug discovery challenges. These include the design of novel targets and therapies; the prediction of safety and toxicity; the stratification of patient populations; and the identification of compounds that increase or decrease protein levels or change the function of a protein.
“Imagine that you could conduct any wet lab experiment that you wanted instantaneously free of charge,” Frey suggests. “Our system allows us to ask questions and get answers up front and turns drug discovery on its head. The predictors tell us if the targets are good and give us a head start in producing highly efficacious and safe steric-blocking oligonucleotides.”
Expediting and improving diagnoses
Next-generation phenotyping (NGP) technologies capture, structure, and analyze human physiological data to produce actionable insights and to facilitate comprehensive, precise genetic evaluations. NGP uses advanced deep-learning algorithms to analyze patients’ photos as well as clinical texts to help clinicians provide a faster and more accurate diagnosis of genetic disorders.
NGP technologies are a specialty of FDNA, a digital health company. It develops AI technologies and software as a service (SaaS) platforms that are used by thousands of clinical and research laboratories globally to aid in the early detection of rare genetic diseases. The company’s database of phenotypic information is exceptionally large, covering more than 5,000 syndromes. All of this information is accessible via cloud computing.
According to Nicole Fleischer, vice president, Product and Collaborations, FDNA, NGP technology, together SaaS and deep learning technology, empower the company’s Face2Gene facial analysis platform.
The platform’s AI algorithms convert a patient photo into deidentified mathematical facial descriptors. After uploading a patient’s portrait photo and adding clinical data, Face2Gene compares the data to thousands of syndromes to quantify similarity, resulting in a prioritized list of syndromes and disease-causing genes.
While used often for children presenting some level of facial dysmorphology, Face2Gene has proven to be useful in other cases. A recent study (Gurovich et al. Nat. Med. 2019; 25: 60–64) found that NGP technologies add significant value in personalized care and may become a standard deep-learning-based genomic tool.
Fleischer indicates that Face2Gene is a pioneer in giving geneticists, pediatric specialists, and laboratory personnel the ability to automatically extract a patient’s phenotype and translate it into a ranked list of genes. “These genes,” she continues, “can then be compared to genetic sequencing results to arrive at possible diagnoses more quickly.”
The company recently launched FDNA Telehealth to enable parents anywhere to quickly access genetic specialists and accurately determine the root cause of genetic diseases. On average, it takes more than 5 years to find a correct diagnosis for a rare disease, and 95% of these diseases lack an FDA-approved treatment. “The wait for a diagnosis and the lack of treatment can be agonizing,” Fleischer states.
Rare diseases, which affect about 500 million people globally, represent an enormous challenge to the healthcare industry—but a significant opportunity, too. Locating each additional patient is critically important not just for that individual, but for every researcher studying the disease, every laboratory developing a test, and every pharmaceutical company developing a drug.

