1. To minimize the potential for sample cross-contamination and nucleic acid carryover from one experiment to the next, use aerosol-resistant pipette tips and designated work areas and pipettes for pre- and post-amplification steps. Wear gloves, and change them often. Do not open reactions after amplification is complete, as opening increases the risk of contaminating subsequent reactions with amplified product. Finally, dUTP can be incorporated into amplification products during PCR to make amplicons susceptible to degradation by uracil N-glycosylase (UNG); this allows you to incorporate UNG into subsequent reactions to control possible carryover contamination.
2. Include an internal standard in your PCRs and RT-PCRs. For example, a second primer pair that amplifies a “housekeeping” gene (i.e., a gene that has constant expression levels among the samples compared) can be included in the reaction (1,2). Amplification of housekeeping genes verifies that the target nucleic acid and reaction components were of acceptable quality and can help account for differences in the amounts of starting nucleic acid. These standards do not account for differences in amplification efficiencies due to differences in product size or primer annealing efficiency between the internal standard and target.
3. Consider whether you want absolute or relative quantitation of your target. Both approaches rely on the quantification cycle (Cq; also known as Ct), which is the cycle number at which the amplification curve crosses the amplification threshold. For absolute quantification, create a standard curve with known amounts of the target template by plotting Cq versus concentration (log concentration), then use the Cq value and an equation derived from linear regression analysis of the standard curve to determine the amount of the unknown sample. For relative quantification, simply compare the Cq values. With an amplification efficiency of 100%, a single template molecule would produce 2n molecules of product after n cycles. Thus, two samples with a Cq difference of 1 have a twofold difference in starting template amount. If the amplification efficiency is less than 100%, the calculation must be adjusted for the decreased efficiency using the following equation:
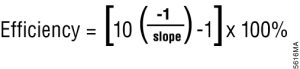
4. When reporting qPCR results, review the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines, as described by Bustin, et al.3 These guidelines outline the minimum information about experimental conditions and PCR parameters required for readers and reviewers to evaluate the qPCR experiment and results and for other scientists to reproduce the results.
5. Like all forms of PCR, qPCR and RT-qPCR requires optimization to ensure sufficient and specific amplification. You may need to optimize PCR parameters such as magnesium concentration, reverse transcription and PCR primer design, template quality, template quantity, cycling parameters, reaction buffer composition and considerations, enzyme concentration and the presence of PCR enhancers and additives.
6. When using fluorescent DNA-binding dyes, perform preliminary experiments to ensure that PCR conditions yield only specific product, as these dyes do not differentiate between specific and nonspecific PCR products. In subsequent reactions, PCR specificity can be verified by melt-curve analysis. Thermal melt curves are generated by allowing all product to form double-stranded DNA at a lower temperature (approximately 60°C) and slowly increasing the temperature to denaturing levels. Product length and sequence affect melting temperature (Tm), so the melt curve reflects amplicon homogeneity. Nonspecific amplification can be identified by broad peaks in the melt curve or peaks with unexpected Tm values. Melt curve analysis is not possible with assays that rely on the 5′→3′ exonuclease activity of Taq DNA polymerase.
7. For probe-based qPCR and RT-qPCR approaches, which use a labeled amplicon-specific probe to produce a fluorescent signal during enzymatic processing of the probe:amplicon hybrid, you should optimize the primer and probe concentrations for each primer/probe combination for optimal results. For gene expression analysis, you also may need to adjust these concentrations based on target abundance. A good starting point is 900 nM for PCR primer concentration and 250 nM for the hydrolysis probe. PCR primer concentration may range from 200 nM to 1 μM, while probe concentration may range from 100 nM to 300 nM.
8. Decide whether one-step RT-PCR or two-step RT-PCR in better for your purposes. For one-step RT-PCR, the RT and PCR steps are performed sequentially in the same tube using the entire amount of cDNA synthesis products as the PCR template. For two-step RT-PCR, RT and PCR steps are performed sequentially, but only a portion of the cDNA products is used as the template for PCR, which is performed in a separate tube. One-step RT-PCR may be more sensitive and requires less pipetting, but two-step RT-PCR allows multiple PCRs from a single RT reaction to quantify multiple targets or perform replicate assays.
9. For RT-qPCR, RNA quality and purity is crucial for success and can affect first-strand cDNA synthesis efficiency. You can use total RNA or poly(A)+ RNA as the template, but both must be intact and free of contaminating genomic DNA. To test for the presence of contaminating genomic DNA in RNA templates, perform a no-reverse transcriptase reaction. Alternatively, design the PCR primers so that they span an intron and genomic DNA is not amplified.
10. A good starting point to detect RNA of an unknown expression level is 100 ng of total RNA template per RT reaction. The amount of RNA required to detect the target of interest depends on the abundance of the target in each sample. A high-copy-number transcript may be detected in as little as 10 pg, while a low-copy-number RNA may require more than 100 ng.



