While most people have heard that at least one of their features—their nose, say, or their eyes—appears to come from one parent or the other, hardly anyone gives any thought to whether individual cells tilt toward mom’s or dad’s genetic contribution. In general, individual cells express both the maternal and the paternal copies, or alleles, of a given gene. Occasionally, however, a cell will arbitrarily begin to use one copy over the other.
This phenomenon—the preferential expression of just one member of a gene pair—is called monoallelic gene expression. It is of interest because it increases variability among cells. What’s more, it likely contributes to phenotypic variance among individuals of identical genotype, including the propensity toward certain diseases.
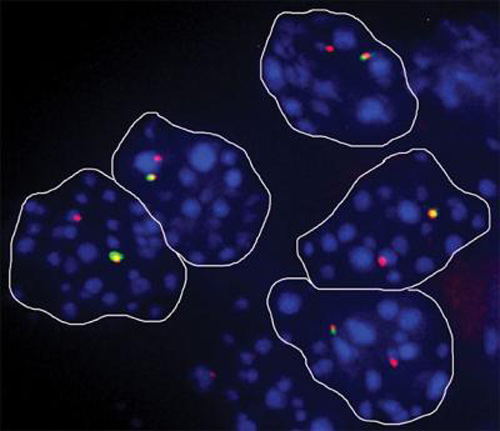
To figure out how monoallelic gene expression arises, scientists have been taking advantage of techniques such as single-cell RNA sequencing. For example, scientists from Cold Spring Harbor Laboratory (CSHL) and the European Molecular Biology Laboratory teamed up to assess the dynamics of monoallelic expression during development through an allele-specific RNA-sequencing screen. They applied this screen to clonal populations of hybrid mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs).
The researchers found that monoallelic gene expression is far more likely to occur in mature, developed cell lines than in their stem cell precursors. They described how they arrived at this result in a paper published February 24 in Developmental Cell (“Random Monoallelic Gene Expression Increases upon Embryonic Stem Cell Differentiation”). They wrote: “We identified 67 and 376 inheritable autosomal random monoallelically expressed genes in ESCs and NPCs, respectively, a 5.6-fold increase upon differentiation.”
“As differentiation occurs, there is a dramatic change in gene expression as a specific program or set of genes is selected to be expressed and a massive reorganization occurs in the nucleus,” said David Spector, Ph.D., a professor at CSHL. “It is these enormous changes that lead to stochastic (that is, variable) monoallelic expression.”
Another interesting result was that expression levels of 8% of the monoallelically expressed genes remained similar between monoallelic and biallelic clones. That is, expression of a gene could be boosted to compensate for what otherwise would be a shortfall.
“This work raises many important questions,” said Spector, “such as: how does the cell know how much of each protein to produce? How much flexibility is there? What is the tipping point toward disease?”
Cells, after all, are exquisitely sensitive to protein amounts. If monoallelic expression were to lead to a shortfall of, say, a tumor suppressor, it could contribute to the development of cancer.