June 15, 2017 (Vol. 37, No. 12)
Rapidly Developing Field Is an Amalgam of Genetics, Automation, and Computer Algorithms
Synthetic biology is an exciting and rapidly evolving field of research that can broadly be defined as the design (or redesign) and construction of novel artificial biological pathways, organisms, or devices. Applying engineering principles to biological components allows us to probe, manipulate, and modify cell function.
The core synthetic biology toolkit is comprised of both biological and engineering functions: molecular manipulation, deep data analysis, and computer algorithms. Below are just a few innovations highlighted at the SynBioBeta conference, which took place in London in April.
“Synthetic Biology is here to stay,” says John Sgouros, consultant, Biomax Informatics. “Customization of molecules and whole microorganisms creates tremendous opportunities in the pharmaceutical industry, environmental biotechnology, and industrial materials. However, optimizing the discovery, genetic engineering, and manufacturing processes driving these innovations is highly dependent on systematic analysis of vast amounts of data.”
Data accumulate rapidly and exponentially, often in an unstructured manner, siloed between internal and external databases, according to Sgouros.
“Biomax’s BioXM knowledge management platform deals with data in a systematic manner: from digitalization to the semantic integration of diverse content and workflows, and further, to systematic feedback of results to find the optimal way forward. Database integration is done by configuration (i.e., point-and-click) without the need to write source code—a huge advantage,” continues Sgouros.
To the end user, the BioXM platform delivers a graphical representation of biomedical knowledge about a given subject, which is easy to explore by clicking on any data field, relationships indicated by connecting arrows. Beneath the user interface is a vast semantically organized knowledge space comprising hundreds of thousands of data elements from published genetic and biochemical information, laboratory notebooks, pharmaceutical databases, electronic health records, and other resources.
Biomax’s proprietary algorithms also extract knowledge from literature and present the end user with a systematic overview of information about genes, biomolecules, drugs, etc., as an interactive map. A powerful example of BioXM application is in personalized medicine, profiling patients with chronic obstructive pulmonary disease (COPD) based on comorbidities and response to rehabilitation. Multidimensional data from over 5,000 COPD patients at the CIRO+ hospital in The Netherlands were integrated with BioXM and analyzed using the Biomax Viscovery software.
The resulting self-organizing maps identified patient clusters with significantly different profiles. Implementation of the clustering in the clinical setting enabled a customized therapeutic approach for each new patient, with considerable costs savings to the hospital. BioXM also forms the knowledge management core for PREPARE [Platform for European Preparedness Against (Re-)emerging Epidemics], analyzing relationships between pathogens and resulting pathophysiology. BioXM is unique as a knowledge management platform seamlessly linking clinical phenotypes to biochemical pathways, pharmacology and genetic variation, along with drug discovery and synthetic biology.

The core synthetic biology toolkit consists of both biological and engineering functions: molecular manipulation and deep data analysis. [iaremenko/Getty Images]
First Synthetic Viruses
“Recognizing natural rules in relationships between DNA, RNA, and transcription machinery is the holy grail of synthetic biology,” says Tahel Altman, M.D., CEO, SynVaccine. “We are continuously perfecting a synthetic biology-driven approach, called SynRad, which systematically evaluates the effect of different nucleic acids features on the resulting “phenotypes” of transcripts and even whole organisms. Once the rules of causality are understood, these rules could be modified to build entirely synthetic organisms with engineered characteristics.”
SynVaccine designs/builds viruses for applications such as human and animal vaccines, specifically aiming at immuno-oncology applications. Dr. Altman explains that modifying viruses by random mutagenesis or by manipulating a specific gene is laborious and time consuming. Synthetic rational design of viruses offers a promise of a continuous pipeline of personalized cancer therapies. SynVaccine can alter viral replication rate, or specificity to a host.
“Most of our rules are proprietary,” contends Dr. Altman. “But exploiting just a few of them underscores our ability to shorten viral vaccine development.”
SynRad algorithms specifically protect desired viral characteristics, such as core protein function or immunogenicity, but introduce hundreds of silent mutation to attenuate viral properties in an optimal way. For example, dengue virus replication depends on the strength of the mRNA folding in different regions. SynVaccine performed a large-scale genomic analysis of reciprocity between dengue RNA folding strength in different positions in the genome and replication rates. The company compared hundreds of individual dengue coding regions to identify signals indicative of functional effect of folding strength. The computationally predicted positions with weak/strong folding signals were modified to create a series of synthetic viruses with variable replication rates. The initial animal data proved that some of the attenuated variants are able to persist in the host without eliciting disease.
Dr. Altman points out that predictable, repeatable, and self-improving generation of synthetic viruses produces a renewed hope for antiviral vaccines as well as creating new hope for cancer treatment using a more accurate viral-based therapy. “Even if we do not understand all the details in the underlying mechanisms, we could discover novel patterns and alter them to change viral specificities,” she says.
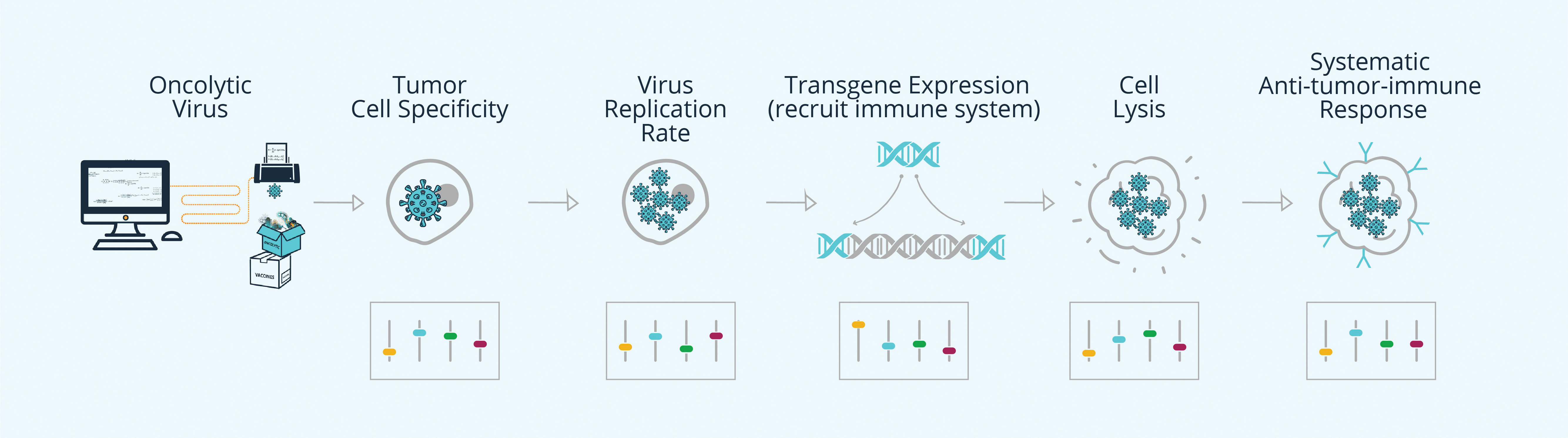
SynVaccine uses a CAD/CAM platform to design, manufacture, and validate viruses, including immuno-oncolytic viruses. The company prepared this figure to depict an optimal immuno-oncolytic virus and its life cycle. Such a virus, the company asserts, must integrate so many specific traits that it could be developed only through a rational design approach.
Biological LEGOs
“Most medicines in the future will be DNA-based biologics,” contends Ryan Cawood, D.Phil., CEO, Oxford Genetics. “As genetic data become more readily available, DNA-based medicines will become indispensable for personalized treatment approaches.” Dr. Cawood described challenges that slow down new biologics R&D: difficulties in discovering new molecules, high complexity of molecular engineering, inefficiency of biologics manufacturing, and the deficiencies of delivery systems. Oxford Genetics guides and supports biologics evolution through understanding how DNA behaves.
“Better DNA in, better biologics out,” offers Dr. Cawood. The foundational technology, SnapFAST, is a LEGO®-like core DNA system of modular blocks. By selecting predesigned DNA parts, such as promotors and enhancers, combined with their own optimized sequence, the company’s customers can make rapid progress in building new human treatments. For example, a novel adenovirus-based gene-therapy system that can deliver DNA into a wide range of cell types, or proprietary algorithms for designing and discovering new DNA sequences and developing antibodies against complex mammalian membrane antigens such as GPCRs.
“Given that we have over 3,000 validated starting DNA systems, this allows us operate in a diverse synthetic biology space, and enables us to offer services using our LEGO-like DNA systems to help discover, develop, and deploy biological therapies more effectively for our commercial collaborators,” says Dr. Cawood.
In 2017, the company also received two substantial grants from Innovate UK for projects that help to integrate the synthetic biology design-build-test cycle to improve protein library design and discovery alongside improving the efficiency of mammalian cell-line engineering. These projects will seek to mine existing datasets, whilst simultaneously developing assays that provide data to allow the development of algorithms that predict cellular behavior and virus and protein production. The company aims to enhance therapeutic molecules along with the bioproduction of the host lines in which they are manufactured.
“We consistently build on previous innovations around DNA design algorithms, automation platforms, and viral packaging systems. The internal R&D projects we are pursuing aim to deliver a quantitative change in the current bioproduction methods, meeting unmet needs of the gene therapy and immunotherapy fields,” says Dr. Cawood.
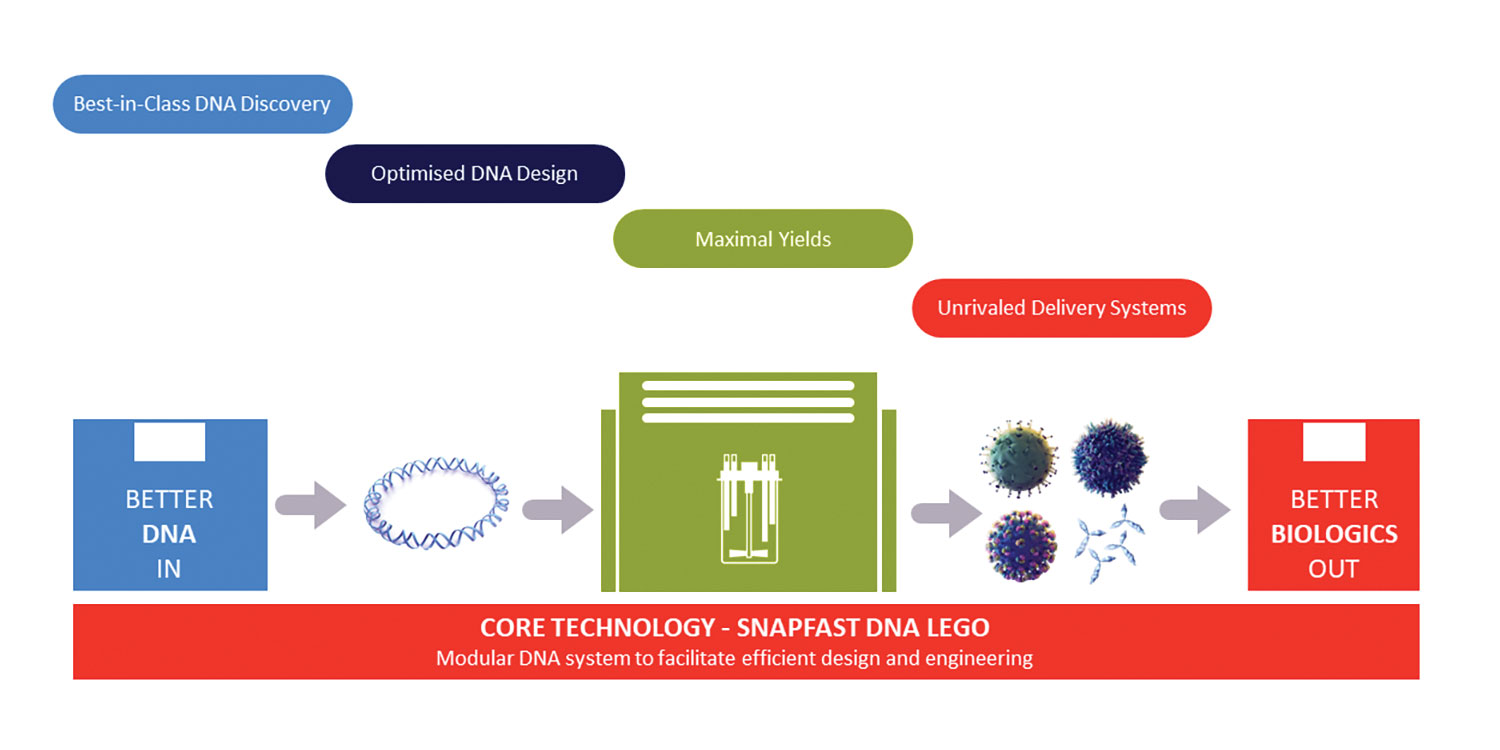
Oxford Genetics’ technologies are based on SnapFast, a modular, LEGO®-like DNA plasmid engineering platform that contains thousands of DNA sections in configurations that allow them to be easily moved from one vector to another. SnapFast is used for efficient DNA design and engineering to address the workflow challenges faced by biotherapeutic companies.
Virtual Cloud Lab
Transcriptic provides access to an on-demand, automated lab in the cloud, obviating the need for laboratory automation, equipment, or even laboratory space itself.
“Moving liquids around by human hands, with or without automation, is the way of the past,” says Yvonne Linney, Ph.D., CEO, Transcriptic.
“Our fully automated lab is available virtually at any time, day or night via our web app. We accelerate knowledge and speed up discovery by providing a seamless connection between experimental design and data. The design of each subsequent experiment is supported by the data from the previous experiment, all within a unified software interface.”
Transcriptic’s robotic cloud lab is a deeply integrated technology stack of biology, robotics, and software made available to its users via the internet. Transcriptic translates a user’s protocol into an open-data standard called Autoprotocol. Predesigned protocols covering a range of typical lab experiments are also available.
Once a Transcriptic user submits an experiment, the robotic cloud lab dynamically provisions enough robotic capability and reagents to complete the experiment at whatever its scale. The underlying robotic infrastructure is a series of Workcells, the size of a shipping containers, filled with fully integrated laboratory equipment. The Workcells form a single virtual lab connected by software all with a single interface to the user. Each Workcell can be programmed separately, and operated remotely.
In a proof-of-concept project (collaboration with Synthego, Transcriptic devised an automated, scalable solution to capture the entire workflow for CRISPR/Cas9-mediated editing, from the synthesis of single-guide RNAs to validation by sequencing of edited mammalian cells. It took less than three weeks to convert the Synthego protocol to Autoprotocol for Transcriptic’s cloud lab.
A collaboration with EpiBiome yielded an automated, high-throughput, and reproducible microbial sample preparation and Ribosomal 16S sequencing pipeline, entirely completed in 7–10 days. Such a rapid and highly scalable protocol offers an opportunity for large-scale microbiome profiling and identification of disease-associated bacteria. “The scientific community struggles with data reproducibility at different locations,” commented Dr. Linney. “In the future, Transcriptic will be able to connect any lab to our cloud, increasing data consistency and transparency.”
Gene Optimization
“Scientists should focus on idea generation, experimental design, and data analysis. We take on the grunt work of synthetic biology: gene synthesis, cloning, and expression,” echoes Axel Trefzer, Ph.D., R&D leader, Thermo Fisher Scientific, adding that GeneArt™ gene synthesis products and services allow for easy design of artificial sequences and their seamless adaptation to a desired host. Users can combine features of different organisms and optimize function without affecting protein function, and thus produce meaningful DNA sequences.
GeneArt gene synthesis supports design of gene sequences of the highest complexity in a range of sizes up to 100 kb. Conversely, GeneArt Strings™ DNA fragments are uncloned DNA fragments up to 3,000 bp in length, assembled from synthetic oligonucleotides.
“Optimization of gene sequences is a key for producing functional DNA in a target organism,” continues Dr. Trefzer. “GeneArt GeneOptimizer™ aims to solve traditional protein expression limitations, such as low yields and unstable, transient expression.”
Using data available from published literature in combination with proprietary data, the GeneOptimizer algorithm determines the optimal gene sequence for a given expression experiment. The company emphasizes high reliability of the design algorithms. The Thermo Fisher Cloud suite of apps interactively guides customers through the design process, highlights problematic areas and enables sequentially optimization until an optimal result is realized. Optimization of genes and DNA fragments have been experimentally proven to increase protein expression rates up to 100-fold in a variety of host systems.
For example, Mapp Biopharmaceutical was able to optimize expression of several monoclonal antibodies, all part of Ebola ZMapp™ therapy. GeneArt Gene Synthesis was also recently utilized in a high-profile application to generate a first-time Zika virus diagnostic assay.
The real-time PCR-based Versant® Zika RNA 1.0 Assay (kPCR) developed by
Siemens Healthineers amplifies and detects all currently identified Zika genotypes, including Asian and African clusters.
“We continue increasing our suite of ‘design-build-test’ apps for scientists” says Oliver Gathmann, manager of bioinformatics, Thermo Fisher Scientific. “This, in combination with our wide set of synthetic biology tools, including DNA libraries, directed evolution, and protein purification services, enable them to execute their vision with shortened timelines and increased reproducibility.
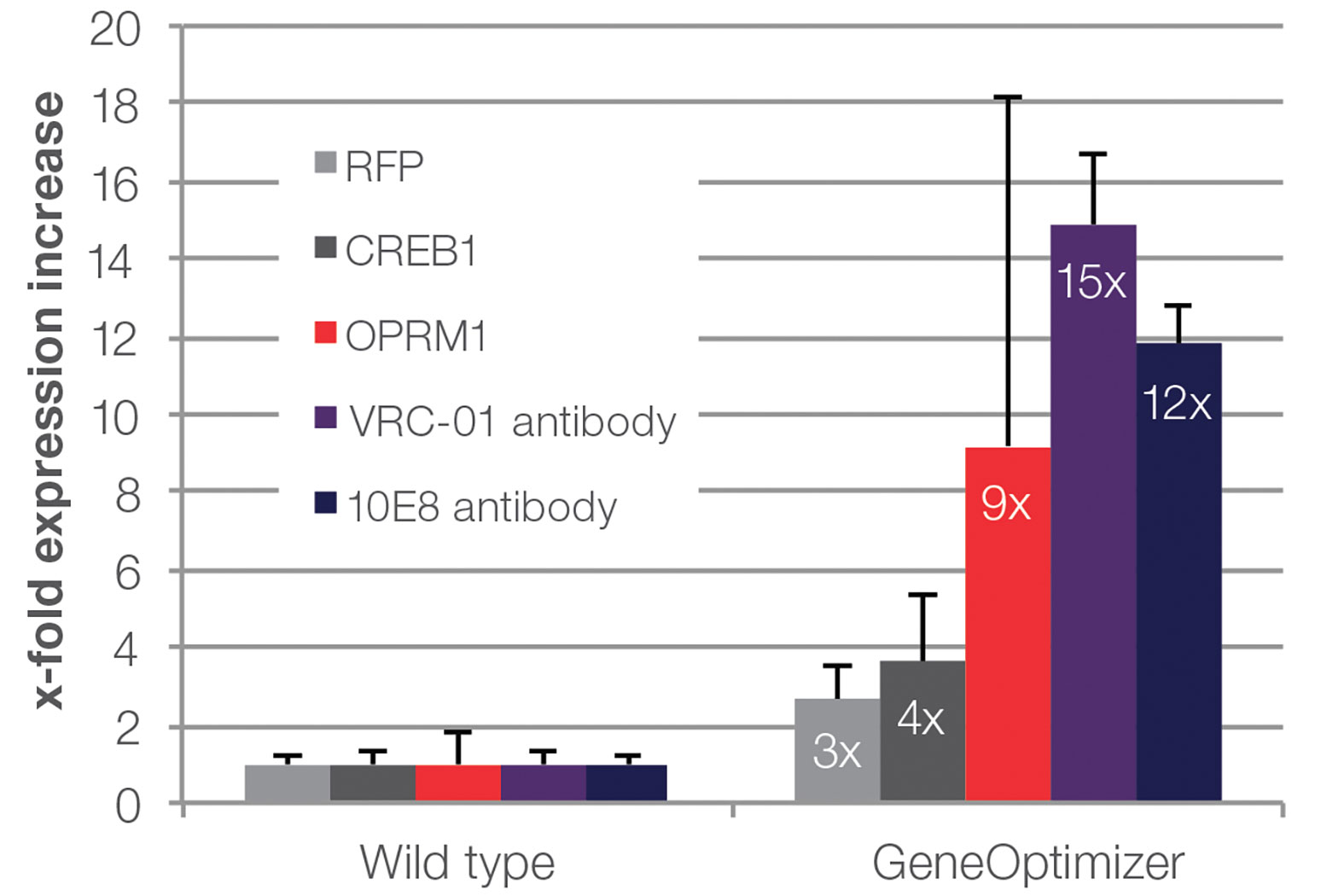
Open reading frames (ORFs) of three intracellular proteins and two antibodies were optimized with Thermo Fisher Scientific’s GeneOptimizer algorithm. Subsequently, protein expression was compared to wild-type counterparts, which was normalized to a value of 1.
Diagnosing Alzheimer’s Using Saliva Biomarkers
Investigators at the Beaumont Research Institute, part of Beaumont Health in Michigan, believe they have discovered small molecules in saliva that will help identify those at risk of developing Alzheimer’s disease. The results of this new study were published recently in the Journal of Alzheimer’s Disease in an article entitled “Diagnostic Biomarkers of Alzheimer’s Disease as Identified in Saliva using 1H NMR-Based Metabolomics.”
Alzheimer’s currently has no cure, few reliable diagnostic tests, and is predicted to reach epidemic proportions worldwide by 2050. As a result, scientists are scrambling to develop methods that can quickly and accurately diagnose the neurodegenerative disorder. In the new study, the Beaumont researchers found that salivary molecules hold promise as reliable diagnostic biomarkers.
“We used metabolomics, a newer technique to study molecules involved in metabolism,” explained senior study investigator Stewart Graham, Ph.D., assistant professor at Oakland University William Beaumont School of Medicine. “Our goal was to find unique patterns of molecules in the saliva of our study participants that could be used to diagnose Alzheimer’s disease in the earliest stages when treatment is considered most effective. Currently, therapies for Alzheimer’s are initiated only after a patient is diagnosed, and treatments offer modest benefits.”
In the current study, 29 adults were divided into three groups: mild cognitive impairment, Alzheimer’s disease, and a control group. After specimens were collected, the researchers positively identified and accurately quantified 57 metabolites. Some of the observed variances in the biomarkers were significant. From their data, the research team was able to make predictions as to those at most risk of developing Alzheimer’s.
“We accurately identified significant concentration changes in 22 metabolites in the saliva of mild cognitive impairment and Alzheimer’s disease patients compared to controls,” the authors wrote. “This pilot study demonstrates the potential for using metabolomics and saliva for the early diagnosis of Alzheimer’s disease.



