January 15, 2016 (Vol. 36, No. 2)
Angelo DePalma Ph.D. Writer GEN
Bioprocessing Assembly Lines Are Being Retooled, Often At the Genomic Scale
Biomanufacturers enjoy a host of tools to optimize the production of therapeutic proteins, including expression systems, media, feeds, and gene-editing tools. Suffice it to say that protein expression is a growth industry.
Industry research firm Future Market Insights (FMI) breaks down the protein expression market into four product areas: competent cells, expression vectors, instruments, and reagents serving demand for research-grade and therapeutic proteins.
FMI has identified noteworthy growth drivers: the rising significance of biologics; innovations in proteomics; and patent expirations among small-molecule drugs. “These demands will boost the overall protein expression market in the coming future,” FMI literature states. “However, [attempts to contain rising costs] in various R&D activities in the fields of biotechnology and pharmaceutical industry as well as market consolidation of a high degree are some restraining factors for this market.”
The largest market for protein expression is expected to emerge in North America, given this region’s “well-established healthcare infrastructure.” North America is followed by Europe, and the Asia-Pacific region shows the highest growth. This information was derived from an FMI report (“Protein Expression Market: Global Industry Analysis and Opportunity Assessment 2015–2025”) that was issued last December.
Despite some bells and whistles, most E. coli production systems have been the same. Now, new systems are being introduced that purport to express proteins more efficiently. [iStock/Scharvik]
Landmark Year
Through the efforts of scientists at Thermo Fisher Scientific, 2015 was a landmark year for transient protein production in CHO cells. The company’s ExpiCHO™ transient expression system achieved multiple g/L levels of protein expression previously thought possible only in stable cell lines, according to Jonathan Zmuda, Ph.D., associate director of cell biology at Thermo Fisher Scientific’s Gibco business unit.
“ExpiCHO allows drug developers to obtain meaningful quantities of protein from CHO cells at the very earliest stages of biologics development,” Dr. Zmuda asserts. “It allows CHO-derived protein to be used from discovery day one through the transition to stable cell lines, bioproduction, clinical trials, and product licensing.”
This has had the effect of streamlining drug development by eliminating the risk of starting a program with HEK 293-derived drug candidates, while also providing an alternative high-expressing system for proteins that are difficult to express in HEK 293.
New E. Coli Expression System
Since E. coli was recruited for service around 1950, hundreds of thousands of publications have sung the praises of this bedrock expression system. But Mehmet Berkmen, Ph.D., staff scientist at New England BioLabs, notes that no more than a dozen distinct protein production strains exist. When production strains are examined closely, all are found to belong to just two basic strains, E. coli K-12 and E. coli B.
“Some strains have ‘bells and whistles,’ but the basic platform is the same,” Dr. Berkmen points out. “People are still looking for engineered lines that express protein more efficiently.”
Most expression systems are based on E. coli B, but that strain is not engineered specifically for protein production. The B strain is somewhat less domesticated than K-12, which has gone through numerous generations of selection for DNA manipulation. “E. coli B is more wild and tends to make protein better,” Dr. Berkmen notes. “But if you ask people why that is the case, they can’t provide an answer.”
New England BioLabs claims that its SHuffle® T7 E. coli expression system represents a breakthrough for microbial fermentation. The bacteria, which are chemically competent E. coli K-12 cells engineered to form proteins containing disulfide bonds in the cytoplasm, are suitable for T7-promoter-driven protein expression. The company has recently produced full-length antibodies, complete with disulfide bonds, in SHuffle organisms, which Dr. Berkmen calls “a significant step toward engineering and developing novel antibody formats and tools.”
New England BioLabs manufactures more than 500 proteins, 98% of them in E. coli. Perhaps even more interesting is the SHuffle system’s ability to express non-disulfide-bonded proteins more efficiently than wild-type E. coli. “SHuffle,” insists Dr. Berkmen, “represents a new chassis for protein production.”
The E. coli bacterium does not form disulfide bonds in its cytoplasm because two reducing pathways maintain the cytoplasmic proteome in its reduced state. Dr. Berkmen’s group knocked out those pathways and inserted a gene for a disulfide bond isomerase that increases fidelity of disulfide bond formation.
In addition to benefits already mentioned, SHuffle has a greatly diminished reducing capacity, permitting the formation of disulfide bonds for proteins that require it for folding and activity. Additionally, the cells, which are under oxidative stress, produce chaperones that also improve folding. For example, the activity of green fluorescent protein (GFP) expressed in SHuffle is much higher than protein produced in wild-type E. coli B.
It should be noted that a lack of glycosylation machinery persists in SHuffle cells. This problem, however, can be circumvented, as demonstrated in a seminal study carried out by Dr. Berkmen and colleagues. This study, which appeared last year in Nature Communications, described how IgG could be produced in SHuffle cells. Specifically, the investigators introduced mutations into the Fc portion of IgG. This resulted in efficient binding of aglycosylated IgG to its cognate receptor FcγRI.
Even in the absence of such ingenuity, E. coli remains a valuable expression system. It can be used to produce diagnostic and reagent proteins, or proteins for which glycosylation is noncritical.
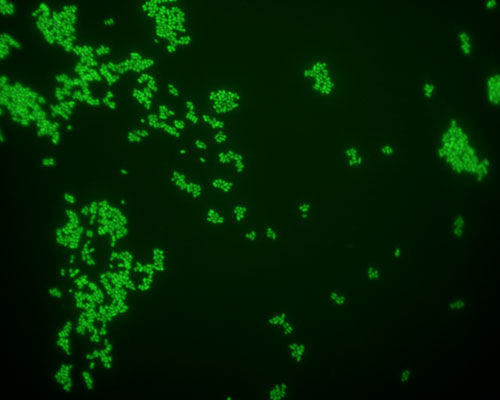
New England BioLabs says that its SHuffle T7 E. coli expression system is able to express non-di-sulfide bonded proteins more efficiently than wild-type E. coli. The actual SHuffle strain expressing GFP is shown here.
“A Matter of Trying”
The principal advantages of using E. coli. are time and cost. “It takes basically one day, more or less, to obtain enough protein to suit many applications,” says David Chereau, Ph.D., CSO at Biozilla, a biotechnology contract research organization. As previously noted, the main disadvantages are lack of glycosylation apparatus and inability to support disulfide bond formation.
Workaround strategies can achieve stable disulfide bonds for some proteins. One strategy involves the following steps: Express the protein as an inclusion body, in insoluble form. Isolate the insoluble fraction. Solubilize this fraction with urea or some other suitable agent. Refold the protein.
“The process is relatively straightforward,” observes Dr. Chereau. “It’s much more difficult to find refolding conditions, which are normally determined empirically.” Refolding requires just the right buffer, salt concentrations, and additives. Also, refolding must be done in an oxidizing environment if disulfide bonds are to be achieved or maintained.
Dr. Chereau is philosophical about CHO cells’ inability to glycosylate: “Lack of glycosylation can be seen as an advantage or an inconvenience, depending.” E. coli is definitely out where glycosylation is a sine qua non. “But for the many applications where glycosylation isn’t needed, E. coli can be advantageous,” comments Dr. Chereau. Diagnostics and reagents are two such products. Additionally, obtaining a crystal structure during protein characterization is easier with glycans absent.
As part of its proof-of-concept services, Biozilla performs rapid screens to determine if E. coli is the right expression system for a particular product. Screening resembles design-of-experiment for mammalian cells, varying plasmids and vectors, as well as expression conditions.
Due to the success of CHO cells, bioprocessors tend to dismiss microbial fermentation, particularly for large proteins. “A lot of people think that expressing large proteins in E coli is difficult,” Dr. Chereau states, “but it’s often just a matter of trying. We have recently expressed a protein of 215 kDa in E. coli, which most people will tell you cannot be done. And we achieved it in very high yield.”
Rapid Prototyping
In June 2015, Invenra, a preclinical stage biotech company specializing in next-generation antibodies and antibody derivatives, entered an agreement with Oxford BioTherapeutics (OBT) to identify and characterize fully human therapeutic monoclonal antibodies (mAbs) against a novel cancer target that OBT has identified.
Invenra’s protein expression platform, through which it is capable of producing hundreds of thousands of full-length antibodies, uses cell-free expression to multiplex up to 10,000 protein variants simultaneously.
“We think of our technology as a rapid prototyping tool for proteins,” says Bryan Glaser, Ph.D., Invenra’s R&D director. “Once we have DNA, we can get protein in less than a day.” Invenra’s expression platform is suitable mainly for discovery and rapid protein prototyping. Yields are quite good: up to 500 μg/mL.
Other firms, such as Sutro Biopharma, are working on cell-free expression at much larger scales. Sutro claims that its Express CF™ technology can produce g/L yields in eight hours.
Cell-free expression involves E. coli extracts, typically S30 (used by most cell-free expression systems) and S12. The numbers reflect centrifugation speed. “Our system is based on S12, which is spun at lower speed than S30,” informs Dr. Glaser. “Our extract also does not undergo dialysis. We think of it as a ‘whole grain’ version.”
In addition to E. coli extracts, additives contain varying quantities of supplemental energy sources, nucleotides, and other small molecules that facilitate in vitro transcription and translation. Every vendor has its own unique blend.
Invenra’s standard mix, which is similar to off-the-shelf products from most commercial sources, is optimized for less complex molecules that don’t require disulfide bonds. Another mix has been optimized to include chaperones formulated to help expression and folding of IgGs and IgG-like molecules.
The upshot: fully functional, correctly folded IgGs and some bispecific antibodies, scFvs, and Fabs. More complex molecules are also possible, but each must be investigated independently. It is possible those could be made, but they would need to be optimized structure by structure. Dr. Glaser says expression capability depends to a large extent on amino acid sequence.
“We can fine-tune expression and folding conditions better than is possible in E. coli,” Dr. Glaser asserts. “We have better control over redox environment to facilitate disulfide bond formation, and we can add chaperones that are not present in E. coli organisms.” Still, the more disulfides the more complex the structure, and the lower the yield.
Dr. Glaser adds that antibody frameworks that express well in E. coli express well in cell-free systems, and ones that don’t express well in bacteria or mammalian cells tend not to express well cell-free. “It could be a framework sequence dependency,” he speculates. “It could be how well that framework folds. Perhaps the best-expressing molecules are those that do not require as much assistance from various chaperones and isomerases.”
Invenra’s expression system lends itself well to large-scale parallelism. The company has developed a credit-card-sized nanowell platform that expresses up to 10,000 unique antibodies per nanowell array. Cell-free expression of IgG using the Invenra nanowell platform system enables the incorporation of functional screening very early into the discovery process.
The ability to screen in excess of 100,000 IgG molecules can reduce the antibody display selection steps and preserve a larger diversity of epitope coverage. In addition, large combinations of binding partners can be empirically tested in various bispecific formats with relevant functional assays to identify the best pair and format for activity.
Getting the Bugs Out
Interest is growing for insect cell expression systems transiently transfected through the baculovirus expression vector system (BEVS). More and more clinical candidates are being generated in insect cells, including development-stage products for respiratory syncytial virus, Ebola virus, and norovirus.
A good deal of BEVS’ success is the ability of insect cells to produce multivalent, multisubunit vaccines through virus-like particles. These proteins can be made at large scale with BEVS for structural studies or to elucidate protein function.
Additionally, insect cells are ideal for making proteins that are toxic to mammalian or E. coli expression systems. BEVS shows its flexibility by providing rapid development cycles for treatments like seasonal influenza or pandemic infection vaccines. Because it is a transient system, BEVS allows for rapid turnaround times compared with mammalian cells, from identification of vaccine candidates to production.







