September 15, 2007 (Vol. 27, No. 16)
Eric J. Devor Ph.D.
Direct Cloning from any Cell or Tissue Source in any Species Is Promoted
In the early 1990s Victor Ambros’ laboratory, then at Harvard University, identified a transcript in Caenorhabditis elegans that contained a 22 nt noncoding RNA that they called lin-4. Ambros’ group discovered that the tiny lin-4 RNA bound to several places in the 3´ untranslated region (UTR) of the heterochronic gene lin-14 and that this served to regulate expression of that gene at the post-transcriptional level.
Calling this noncoding species a microRNA, Ambros’ discovery opened the door on a previously unsuspected world of small, noncoding, regulatory RNAs that, today, has grown to thousands of transcription units encompassing six distinct families of which at least one has been found in nearly every eukaryotic species examined.
Small RNAs are crucial elements in a host of cellular processes including development, apoptosis, genome organization, and several diseases, notably cancer. For this reason, numerous products, protocols, and software packages have been developed to aid in small RNA-expression profiling and target prediction.
However, little has been done in the way of aiding primary small RNA discovery. Only miRNAs display the thermodynamically stable hairpin structures that aid in in silico discovery and even this is limited to sequenced genomes. To address the need to be able to identify small RNAs in vitro in any species and in any tissue, Integrated DNA Technologies (IDT; www.idtdna.com) has developed a line of small RNA cloning aids. The centerpiece of this line is the miRCat™ small RNA cloning kit.
miRCat Small RNA Cloning
A number of methods have been proposed for direct in vitro cloning of small RNAs. We have found that the most efficient method involves the use of a preactivated, adenylated 3´-cloning linker and a compatible 5´-linker.
IDT offers three adenylated 3´-linkers with different sequences/cloning sites (Linker 1, Linker 2, and Linker 3) and a 5´-linker that is compatible with all three of the 3´-linkers (5´ M.R.S. Linker).
In July, IDT introduced a new kit to help with all aspects of the miRNA cloning process that pairs Linker 1 with the 5´ M.R.S. Linker. Called miRCat (for miRNA concatemer cloning), this kit permits direct cloning of small RNAs from any cell or tissue source in any species. The kit includes PCR and reverse transcription primers, T4 RNA and DNA Ligase enzymes, and other necessary buffers, reagents, and optimized protocols.
Protocol Overview
The complete cloning strategy is shown in Figure 1. Briefly, total cellular RNA is isolated using an organic extraction method (such as Trizol, RNA STAT-60, or equivalent). The small RNA population is isolated from a denaturing polyacrylamide gel using a synthetic 21 mer internal RNA control (miSPIKE™, Figure 2).
The purified small RNAs, including the spiked in 21 mer internal control, are 3´ ligated with a preactivated, adenylated linker (rAppCTGTAGGCACCATCAATddC) using T4 RNA Ligase in the absence of ATP. Linkered RNAs are then purified from a second denaturing polyacrylamide gel. These purified RNAs are next 5´ linkered with the DNA/RNA chimeric 5´ M.R.S. Linker (TGGAATrUrCrUrCrGrGrGrCrArCrCrArArGrGrU) using T4 RNA Ligase, this time in the presence of 10 mM ATP.
The miSPIKE internal control is made without a 5´ phosphate and, therefore, is unable to accept the 5´ M.R.S. Linker. Thus, the doubly ligated species will derive from the original tissue RNA purification and not the miSPIKE marker.
These species are reverse transcribed and then PCR amplified using the RT/REV primer 5´-GATTGATGGTGCCTACAG-3´ and the FOR primer 5´- TGGAATTCTCGGGCACC-3´. PCR amplification conditions are 95ºC5:00; (95ºC0:30; 52ºC0:30; 72ºC0:30) x 35 cycles; 72ºC7:00. The expected amplicon size is ~70 bp.
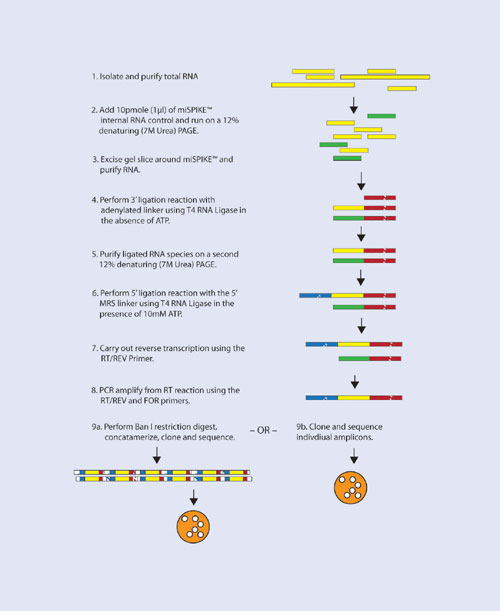
Figure 1
Cloning Options
Two cloning options are available. Amplicons may be cloned directly into a PCR cloning vector (such as pGEM T-EASY, TOPO-TA, or equivalent). Alternatively, amplicons can be digested with Ban I restriction endonuclease, concatemerized using T4 DNA Ligase, and cloned into a suitable PCR cloning vector.
The miRCat kit is supported by several additional products including reagents that enable a 5´ ligation independent small RNA cloning method (miRCat-33™), a 31 mer internal RNA (piSPIKE™) to facilitate cloning piRNAs, and two 454 PCR primers that will modify the small RNA libraries into a form that is appropriately tagged for deep sequencing using the Roche/454 GS FLX or GS20 systems (454 Primer Sets I and II).
The discovery of new members of the extant families of small, noncoding, regulatory RNAs and of additional families will be greatly facilitated by the availability of a reliable, generic method for cloning these small RNAs directly from cellular RNAs of any tissue and any species. The new line of small RNA cloning aids from Integrated DNA Technologies is specifically designed to meet this need.
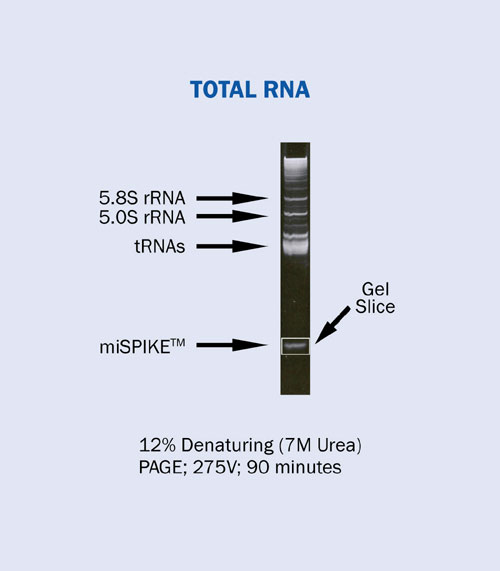
Figure 2
Eric J. Devor, Ph.D., is a senior research scientist at IDT. Web: www.idtdna.com. Phone: (319) 626-8450.
E-mail: [email protected].







