March 15, 2017 (Vol. 37, No. 6)
Philippe Desjardins Scientific Marketing Consultant Venture Novo, LLC
Nucleic Acid Analytes Are Reliable Sources of Data for Diagnostic Calculations
Advances in microanalytic methodologies, instrumentation sensitivity, and computational power have enabled the use of fleetingly small amounts of analyte to further stratify individualized molecular information in real time, potentially revolutionizing molecular diagnostics.
For example, circulating cell-free DNA has become a promising avenue for molecular diagnostics. The shedding of cellular material into the bloodstream provides a constant source of analyte that can be acquired directly via liquid biospies.
Scientists are also turning to nucleic acid analytes beyond genomic DNA for diagnostic purposes. As cells continually transcribe and translate genetic information, the RNA intermediary may be a means of providing profound new insights into the health of any given tissue.
RNA Expression
“One of the advantages of looking at RNA expression, whether it be in tissues, or cells, or even organs, is that it gives a snapshot of the immediate state of that individual patient, which in turn is driven by multiple factors such as environment, stress, and of course disease,” states Jon Armstrong, chief science officer, Cofactor Genomics. “What is exciting about RNA is that expression will go up or down depending on the state of each tissue. We think of RNA as a platform. It’s obvious to us that RNA will become prime time—as prime time as DNA, if not more so—as a tool for diagnostics.”
At any given moment in every tissue, there is a vast array of RNA expression controlling a complex interacting network of cellular processes.
“We think in terms of those many points of expression being similar to looking at a starry sky,” Armstrong elaborates. “There is a unique constellation of expression signals for every tumor, tissue, or formalin-fixed paraffin-embedded (FFPE) sample. The information generated from a cancer sample is compared against expression signals of potentially hundreds of samples of the same cancer type, uncovering expression patterns that make the sample unique.”
This idea, which could be called constellation diagnostics, goes beyond the usual comparative expression test between tumor cells and normal cells. When a cancer signature (or pattern of RNA expression) is obtained from a particular sample and compared to all other cancer signatures, there is no need for a “normal” signature.
It becomes possible to recognize what makes a specific cancer sample unique, not only with respect to a normal sample, but with respect to any other cancer sample. That is, any cancer type may be distinguished from and other cancer type for which RNA expression data is available. This approach, Armstrong suggests, “takes us one step closer to having a better snapshot of the actual patient.”
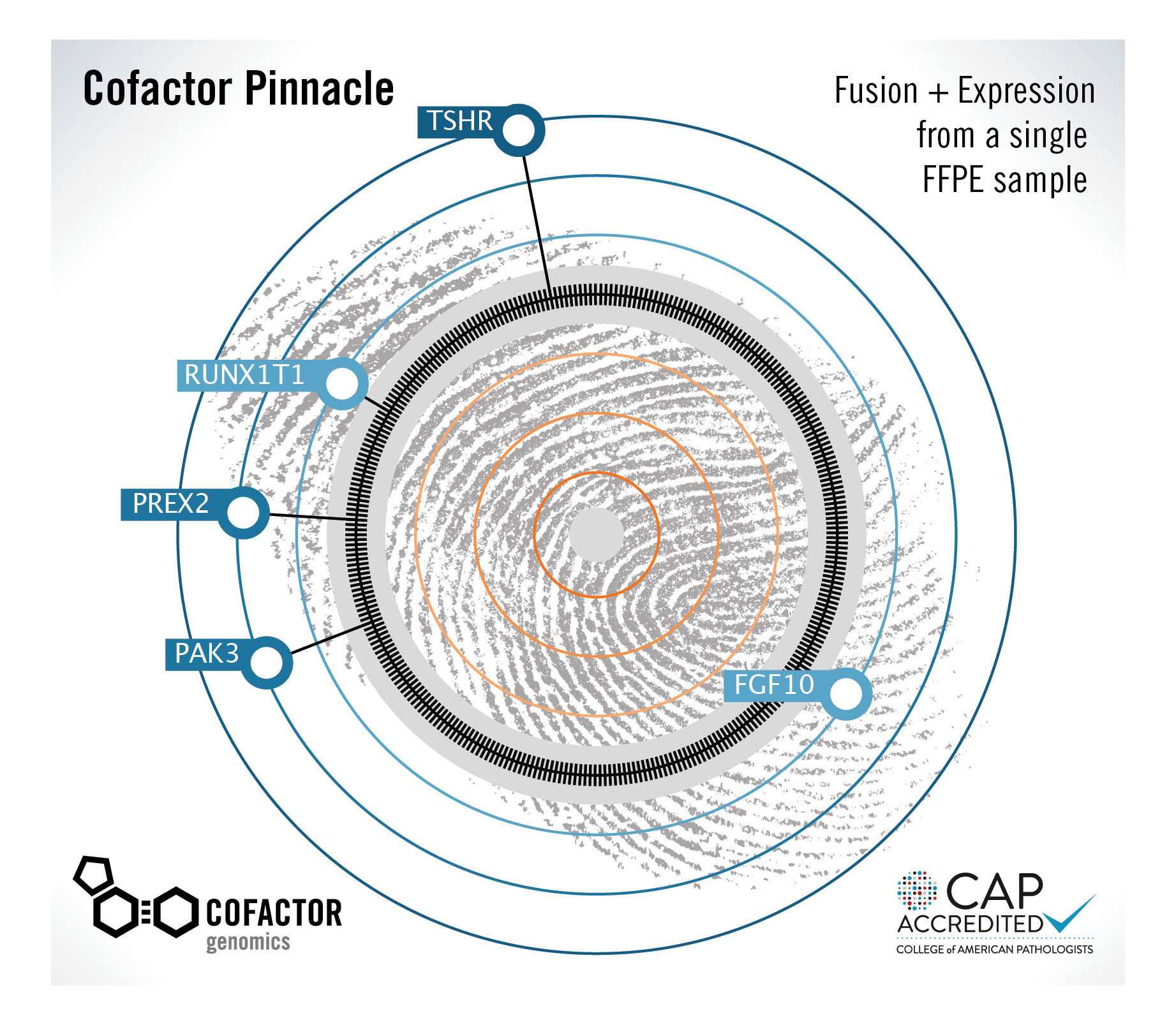
Cofactor Genomics’ Pinnacle expression plot is derived by comparing the RNA “fingerprint” of a cancer sample to hundreds of other samples of the same cancer type. By displaying aberrant genes that make a patient’s sample unique, the plot could lead to better treatment decisions.
Liquid Biopsies
“We believe liquid biopsies are the missing cornerstone to personalized medicine,” says Christian Jurinke, Ph.D., managing director, Stratec Molecular. “We now have the opportunity to use a noninvasive approach with just a simple blood draw, instead of piercing an organ and removing a bit of tissue.”
Liquid biospies, Dr. Jurinke notes, promise to advance oncology and are already having an impact in other areas. “Prenatal diagnostics has been revolutionized by analyzing trace amounts of fetal DNA in maternal plasma,” he points out. “The introduction of noninvasive diagnostics has had a great impact on the entire field.”
The noninvasive nature of liquid biopsies, comments Dr. Jurinke, affords the following advantages:
• Patients naturally prefer noninvasive methods over more invasive diagnostic procedures such as surgery.
• Complications from liquid biospy analysis are nearly nonexistent, compared to those that can arise from traditional procedures such as direct lung cancer biopsies.
• Monitoring the progress of cancer therapy can be performed much more frequently due to the minimal impact of this approach, providing a better opportunity to change treatment and affect prognosis.
“For some patients, frequency matters a lot,” Dr. Jurinke insists. “For example, breast cancer has a high rate of recurrence. Having the ability to monitor progress more frequently, and potentially detect recurrence very early on, significantly increases the chances of survival.”
Is it possible that liquid biopsies have advantages besides noninvasiveness over traditional tumor biopsies? This question drew from Dr. Jurinke a nuanced response: “Liquid biopsies may potentially contain additional information about metastasis from distant sites, as opposed to information attained only from direct targeting of the tumor. Cell-free DNA, however, is a much more challenging target. It is harder to analyze than genomic DNA taken directly from tissue.
“The half-life of circulating DNA in blood is very short, and the amount of cell-free DNA from cancers is extremely low. Although these are significant challenges, they can be overcome by taking a lot of care during preanalytics and the entire workflow, as well as having very sensitive analysis methods.”

The InviGenius Plus by Stratec Molecular is an automated device that can extract cell-free DNA from up to 4 mL of plasma. The large sample capacity enables extraction of more cell-free DNA, which can then be analyzed downstream, where detection methods such as real-time PCR or next-generation sequencing can be used.
Assessments of Trauma and Transplantation
Cell-free DNA is not only enriching cancer and prenatal diagnosis, it is also being investigated as a potential analyte for trauma and organ transplantation. To explain the rationale behind the use of cell-free DNA in trauma patients, Dana W.Y. Tsui, Ph.D., attending geneticist, Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, revisits the idea that circulating DNA in the bloodstream may be a byproduct of apoptosis.
“Basically, when cells die, they release cell-free DNA into the circulation,” Dr. Tsui explains. “Although very exploratory in terms of potential clinical utility, using this analyte to measure the extent of cell death could correlate to the seriousness of traumatic injury.”
Cell-free DNA, Dr. Tsui asserts, may also be useful in organ transplantation: “If you think about it, organ transplantation between donor and recipient is essentially a combination of genomes—two different genomes in one body. Monitoring levels of circulating DNA shed from the donated organ may provide an early indication of apoptosis.
“The beauty of cell-free DNA is that it tells us something about the dynamic activity of the donated organ. Cell-free DNA may serve as a noninvasive marker to pick up potential rejection in advance.”
Furthermore, any changes in the activity of the donated organ will be in real-time, due to the relatively rapid clearance of cell-free DNA from the circulation.
“This is a very exciting time for using cell-free DNA as a tool for molecular diagnostics,” Dr. Tsui remarks. “There is a lot of interest in this area as more sensitive technologies become available that can tease out this type of information.
“Cell-free DNA applications are so tightly linked that knowledge learned from one application helps in the development of the others. For example, cancer management has been greatly accelerated by the development of noninvasive testing; so by extension, transplantation and trauma applications will also benefit in parallel.”
Reference Standards
Fundamental shifts in diagnostic technologies have also significantly influenced the development of reference standards that can accurately reflect the true nature of a sample, such as the complexities found in cancer.
“We’re increasingly looking at many biomarkers in parallel,” says Brian Burke, Ph.D., director of business development, Horizon Discovery, Cambridge, U.K. “This approach helps us understand the heterogeneity of cancer. We’re starting to use multiple analytes—such as solid tumor material, circulating tumor cells, and circulating free DNA—to build a picture of what is going on in the patient.
“Our reference standards are based on engineering a cancer mutation into a cell line and then having a matched normal cell line without the mutation. Through a combination of different cell lines, we are able recapitulate something that mimics the actual tumor mix with the corresponding ratios of normal genes to nucleotide polymorphisms and other types of genetic variations that drive cancer.”
The reference standards can be used to validate biomarker assays. Also, these standards contribute to platform development, helping answer questions regarding sensitivity, reproducibility, and specificity.
“As we move from solid tumor specimens to smaller amounts of cancer in liquid biopsies,” informs Dr. Burke, “we confront the challenge of looking for vanishingly small numbers of molecules in a very complex mixture of blood.”
The development of highly sensitive and reliable detection methods is facilitated if suitable reference standards are available. “The ideal scenario,” Dr. Burke concludes, “is to develop a universal reference standard that can cover multiple analytes and accurately represent the full complexity of a disease state.”
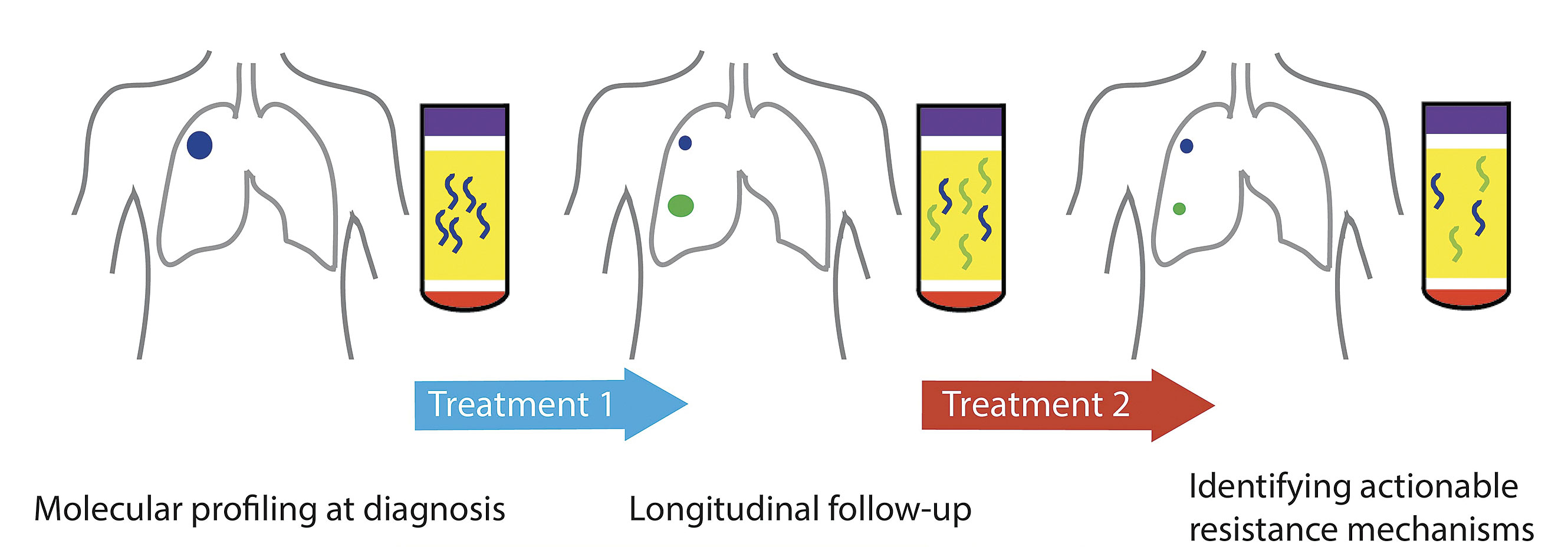
Circulating tumor nucleic acids present opportunities for noninvasive cancer management. Because these nucleic acids may be obtained safely and easily, they facilitate serial monitoring, which may encompass treatment follow-up and detect nascent resistance. [Memorial Sloan Kettering Cancer Center]
Harmonizing Companion Diagnostics
The idea of a universal standard naturally lends itself to the concept of harmonization between various diagnostic tests. Companion diagnostics has predominately been developed with the view of one diagnostic test supporting one drug to determine therapeutic applicability for one patient.
“When there was a limited number of drugs directed against the same target, harmonization was a relatively straightforward concept,” recalls Scott D. Patterson, Ph.D., vice president, biomarker sciences, Gilead Sciences. “It was, in any case, necessary to bring the idea of diagnostics supporting therapeutics into play.”
“However, when multiple drugs are being developed at a similar but asynchronous time frame, each with its own diagnostic, harmonization is a huge challenge,” Dr. Patterson continues. “If all of those drug and diagnostic combinations gain approval, the result is multiple diagnostic tests for different drugs targeting the same biomarker.”
One good example is PD-L1 expression testing for immuno-oncology applications.
“There are multiple diagnostics and drugs available, some of which are companion diagnostics, and some of which are complementary diagnostics. So now we have multiple different tests for the same purpose,” Dr. Patterson adds. “You want competition, but you don’t want confusion.”
One solution is to develop a diagnostic test that encompasses a broad panel of targets and enables simultaneous testing of a patient for different therapies. “In other words, the goal is to develop a testing panel that could support multiple drugs,” explains Dr. Patterson. “That is where the field is trying to go at the moment.”
“We are always looking at the risk-benefit ratio for any therapeutic decision,” he stresses. “There are some biomarkers that are positive selection biomarkers, and there are others that are negative selection biomarkers. It would be wonderful if there were analytically validated panels that drug developers could use to harmonize different tests and identify patients who could benefit most from specific therapies.”

Horizon Discovery provides genetically defined, human genomic reference standards that can help laboratories ensure the sensitivity, specificity, and accuracy of their assays and workflows. The company’s HDx products include cell-free DNA reference standards.
High-Throughput Molecular Clinical Research via LC/MS
Hemoglobin profiling is an essential technique for a number of clinical research applications, including hemoglobinopathy workflows for the identification of genetic blood disorders such as sickle-cell disease. The genetic defects associated with these diseases result in the formation of abnormal globin chains in the hemoglobin molecule, caused by sequence truncations, omissions, and SNPs during protein translation.
Blood profiling analysis must be capable of confident characterization and relative quantitation of both normal and abnormal hemoglobin variants.
With over 1,000 forms of hemoglobin reported to date, traditional targeted research screening approaches are unable to comprehensively analyze all molecules of interest on a convenient timescale, according to Scott Peterman, Ph.D., marketing manager, mass spectrometry, biomarker research, Thermo Fisher Scientific.
“Furthermore, with so many samples to screen—each one as individual and valuable as the next, it is critical that workflows are reliable, robust, and deliver accurate results each and every time,” adds Dr. Peterman.
Research Study
In a recent research study, a high-throughput hemoglobinopathy routine was developed to quantify normal alpha and beta human hemoglobin chains, alongside abnormal variants, in complex mixtures containing human and bovine hemoglobin. The Thermo Scientific Vanquish* Ultra High Performance Liquid Chromatography (UHPLC) system was used to deliver “superior,” in the words of Dr. Peterman, separation power, enabling confident global protein profiling regardless of sample complexity.
“When used in conjunction with the advanced resolution and dynamic range of the Thermo Scientific Q Exactive Focus* mass spectrometer, the method demonstrated exceptional reproducibility for fast and reliable large-scale analysis,” he continues.
Automated high-throughput workflows capable of handling large numbers of complex samples is essential. The latest advances in UHPLC and mass spectrometer technology are helping researchers understand how they might analyze complex biological samples more rapidly, robustly, and with greater confidence in their results, says Dr. Peterman.
* For Research Use Only. Not for use in diagnostic procedures.







