April 15, 2014 (Vol. 34, No. 8)
Generation of Monoclonal Antibodies against Difficult Membrane Protein Targets
Biologics are the fastest growing class of pharmaceuticals, with hundreds of monoclonal antibodies (mAbs) currently being developed and over thirty mAbs approved for the treatment of conditions ranging from cancer to autoimmune and inflammatory diseases. In the case of membrane protein targets, mAbs can avoid many of the pitfalls of small molecule drugs such as poor bioavailability and hydrophobicity that result from targeting protein regions embedded in a lipid bilayer. However, mAb discovery efforts against membrane proteins have been hindered by the difficulties of working with these complex targets.
Integral membrane proteins such as G protein-coupled receptors (GPCRs), ion channels, and transporters span the lipid bilayer multiple times and mediate many of the cell’s critical signaling and transport functions. Standard detergent-based protein purification techniques are often used to extract such proteins for use as antigens in mAb discovery, but can alter the structure of embedded proteins that depend on the lipid bilayer for correct folding and functioning. As a result, mAb generation against these complex targets requires alternative strategies. Whole cells and peptide immunogens have been used successfully for some membrane proteins, but these approaches have yielded very few conformational mAbs suitable for therapeutics. Specialized techniques such as nanodiscs, stabilized membrane proteins, and reconstituted liposomes have been pursued to generate mAbs against correctly folded membrane proteins, but results have been limited primarily to well-expressed membrane proteins.
Because of the limitations of current techniques, there are fewer than a dozen inhibitory mAbs against the approximately 400 transporters and 400 ion channels in the human genome, and the vast majority of the additional 1,600 integral membrane proteins also lack inhibitory mAbs. To date, the tetra-spanning B-cell antigen CD20 is the only multi-pass membrane protein targeted by FDA-approved mAbs.
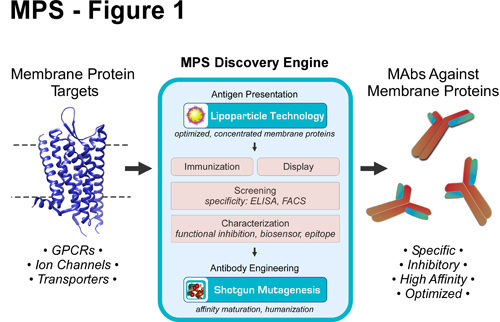
The MPS Discovery Engine
To systematically isolate, optimize, and characterize mAbs against difficult membrane protein targets, Integral Molecular has developed the MPS Discovery Engine (Figure 1), which overcomes many of the challenges associated with membrane proteins. The platform uses Lipoparticles containing concentrated membrane proteins to isolate conformationally sensitive, inhibitory mAbs against native protein targets. MAbs are characterized for specificity, binding kinetics, epitope location, and functional efficacy, enabling the identification of promising lead candidates. mAbs are optimized as needed using Shotgun Mutagenesis, a comprehensive mutagenesis platform for structurally complex human proteins, to affinity mature or humanize mAbs for preclinical development. MPS has been used to derive mAbs against integral membrane proteins that have never previously been successfully targeted, including therapeutically relevant GPCRs, ion channels, and transporters.
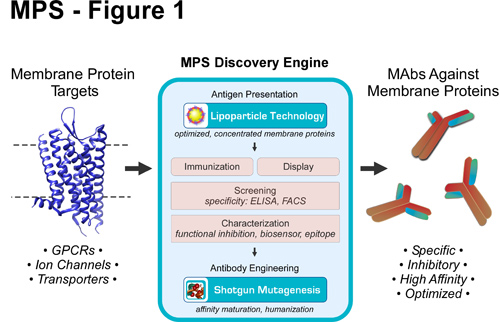
Figure 1. MPS Discovery Engine for the discovery of mAbs against membrane protein targets. MPS uses Lipoparticle and Shotgun Mutagenesis technologies to generate conformation-dependent mAbs against difficult membrane proteins. Both Lipoparticle immunization and display technologies are used, depending on the target, to isolate mAbs, which are then further characterized and optimized as necessary.
A primary bottleneck in membrane protein mAb discovery is obtaining sufficient quantities of correctly folded target proteins for immunization and screening. MPS addresses this obstacle by concentrating structurally intact membrane proteins in Lipoparticles, which are virus-like particles (VLPs) that display target proteins in their native conformation at levels 10- to 100-fold higher than on cells or in membrane preparations. To further boost expression of the most difficult proteins, constructs are engineered to maximize transcription, protein folding, stability, and transit to the cell surface. For example, protein engineering has been essential in enabling high expression of KV1.3, NaV1.7, and TRP ion channels (Figure 2A), which are targets for autoimmune and pain disorders. These proteins are notoriously difficult to work with due to their low surface expression and cytotoxicity, but modifications such as mutations, truncations, and chimeras enabled 15 to 50-fold higher expression on Lipoparticles.
Membrane proteins are often highly conserved and typically present relatively small extracellular regions to the immune system. Hence, limited animal immunogenicity is often a stumbling block for mAb discovery. To elicit more robust immune responses, divergent species such as chickens, llamas, and sharks have been used in Lipoparticle immunization projects, in addition to traditional mice and rabbits. MPS has been used to target all major classes of membrane proteins including GPCRs, ion channels, transporters, tetraspanners, oligomeric proteins, and viral envelope proteins and has successfully produced high-titer immune responses in diverse animal hosts (Figure 2B).
As an example, the glucose transporter GLUT4 is a complex membrane protein comprised of twelve transmembrane helices linked by very small, highly conserved extracellular loops. Due to this challenging structure, mAbs that can recognize glucose transporters on the cell surface had not been previously isolated using standard techniques. With MPS, GLUT4 Lipoparticles were used to immunize chickens, yielding highly reactive antibodies that were isolated by Lipoparticle-phage display (Figure 2C). Despite the complexities of the glucose transporter target, this campaign enabled the first-ever isolation of mAbs that can specifically recognize this family of membrane proteins on cells.

Figure 2. MPS enables the discovery of antibodies against diverse membrane protein targets. (A) The ion channels NaV1.7, KV1.3, and TRP were engineered to enable high levels of expression 15–50 times their wild type levels. (B) Immunizations with KV1.3, GLUT4, and the GPCRs Galanin receptor 1 (GALR1) and Glucagon receptor (GCGR) generated highly reactive sera specific to each target. (C) Phage display using GLUT4 Lipoparticles yielded a large number of antibodies with high selectivity for their target protein. (D) MAbs against P2X3, derived using Lipoparticles, functionally inhibited the ion channel and reduced ligand-triggered calcium flux in human cells. (E) P2X3-specific mAbs bind their target protein with subnanomolar affinity, as determined by biosensor analysis using P2X3 Lipoparticles. (F) Epitope mapping of CXCR2 and CXCR4 mAbs by Shotgun Mutagenesis revealed interactions with conformational epitopes and informs the mechanism of action of each mAb.
Inhibitory mAbs against Membrane Proteins
MAb discovery against membrane proteins also faces the challenge of isolating conformation-dependent mAbs, which are the most likely to exert a functional effect with therapeutic potential. Screening of mAb clones against Lipoparticles allows conformational mAbs to be identified since Lipoparticles display natively folded membrane proteins captured directly from the cell surface in their correct orientation. For example, nearly thirty unique mAbs against the ion channel P2X3, a validated therapeutic target for pain, were isolated using Lipoparticles. Thorough characterization of these mAbs revealed that they bind distinct conformational epitopes on P2X3 with high specificity and subnanomolar affinity. Many of these mAbs were found to inhibit the transduction of ATP signaling in vitro (Figure 2D), as well as electrical currents in primary sensory neurons ex vivo, thus representing some of the few inhibitory mAbs against any human ion channel.
Additional studies of mAb affinity and mechanism of action are important for selecting lead candidates with the most therapeutic promise, but for membrane proteins these characteristics are often difficult to evaluate using conventional techniques. With MPS, detailed mAb binding kinetics are assessed by optical biosensor using immobilized Lipoparticles enriched for the target protein (Figure 2E). The specific residues that constitute the mAb epitope can be identified by Shotgun Mutagenesis (Figure 2F), informing the mechanism of action of the antibody as well as enabling intellectual property protection. Finally, a number of mAb features can be engineered using Shotgun Mutagenesis, such as maximal (germline) humanization and affinity maturation.
The discovery of mAbs targeting integral membrane proteins using traditional approaches has been challenging to date. The MPS Discovery Engine is a novel platform that provides access to new mAb targets, helping to bring first-in-class mAb therapeutics to the market. This approach provides a pathway for the generation of unique, highly specific, functional mAbs against previously intractable membrane proteins.
Virginie S. Adam, Ph.D. and Soma S. R. Banik, Ph.D. are research and communications scientists, and Benjamin J. Doranz, Ph.D. ([email protected]), is president and CSO at Integral Molecular.