November 15, 2017 (Vol. 37, No. 20)
A Strategy for Projects Requiring Complex Genetic Modifications
Pressure to accelerate drug discovery is high, and can be impacted by how mouse models are generated and bred. This tutorial reviews the use of two techniques that, when combined, reduce the time needed to reach the experimental cohort stage when CRISPR/Cas9-mediated gene editing in embryos is not the best option.
Mouse models are often customized to suit study objectives and improve translatability of results. Yet the process to generate a new model and produce a suitably sized cohort can be lengthy, especially when complex genetic modifications are required. While the fastest gene-editing protocols—such as CRISPR/Cas9 in embryos—are efficient when used for simple modifications, complex modifications are typically generated more efficiently using embryonic stem (ES) cell–mediated techniques. The timeline for this approach depends on factors such as the number of animals generated in Phase I, cohort size, and genotype. However, the average time for genome modification, generation of chimeras, and production of a few heterozygous mice is 42 weeks. An additional 28 weeks of breeding usually is needed to produce a study-size cohort, for a total of 70 weeks.
For research projects that are best accomplished using an ES cell-mediated approach, molecular analysis of sperm to directly determine germline chimerism can be combined with in vitro fertilization (IVF) to speed cohort production. This strategy reduces the model generation timeline by 12–16 weeks, without sacrificing quality control (see Figure 1).
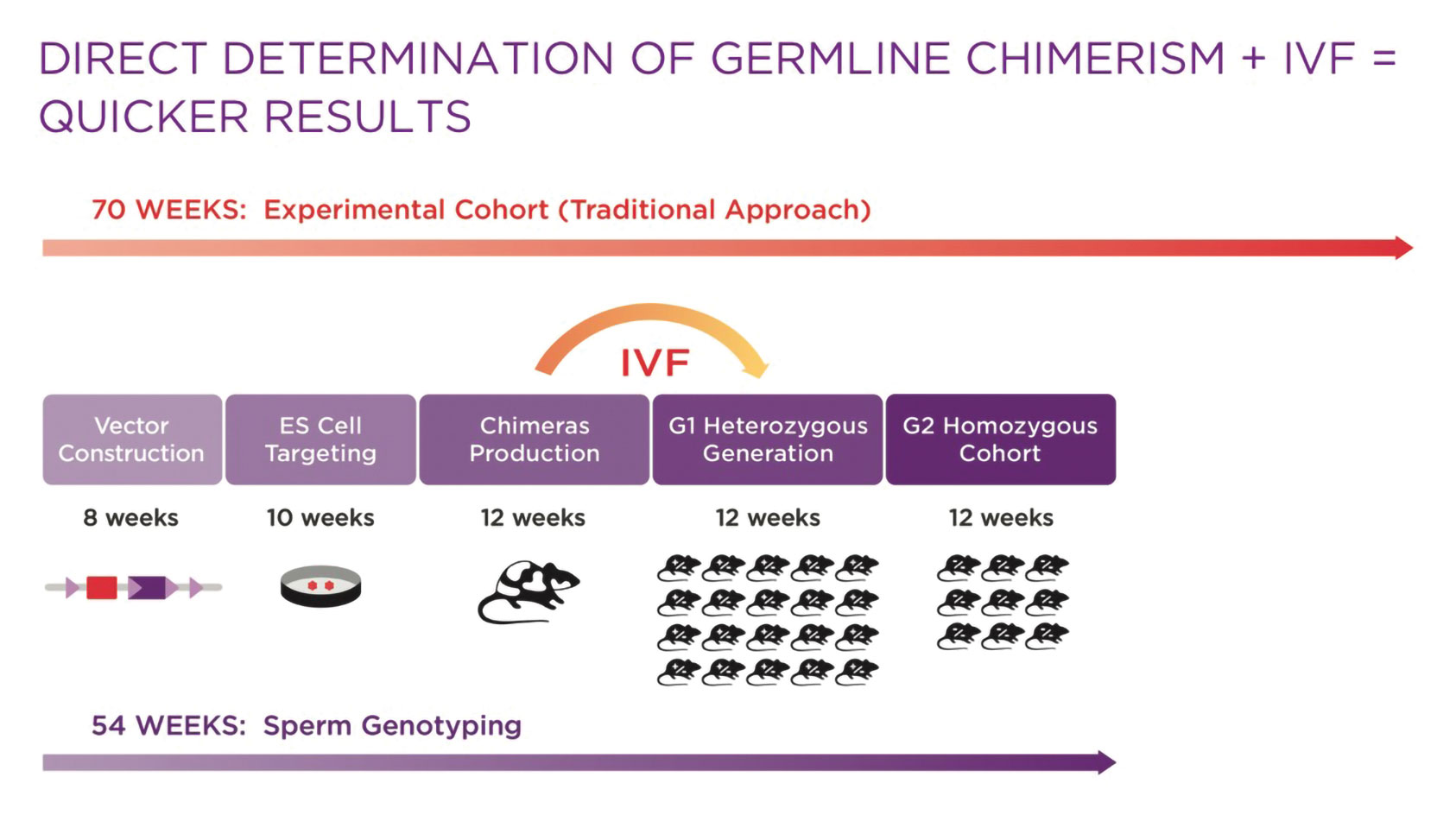
Figure 1. Using an embryonic stem (ES) cell-mediated approach to mouse model generation reduces the timeline by 12–16 weeks, without sacrificing quality control.
Employing Molecular Analysis of Sperm to Directly Determine Germline Chimerism
Identifying the male founder mice that are most likely to quickly transmit the modified allele accelerates the initial breeding steps. Molecular analysis of sperm from male chimeras can be used to make this identification.
When using ES cell-based methods for genome modification, the ES cells are first manipulated in culture and then injected into blastocyst-stage embryos. The intent is that they will integrate into the developing embryo and become germ cells, enabling transmission of the genetic modification to the next generation. The injected embryos become chimeras—mice with multiple, genetically distinct cell lineages. While there are techniques to generate founder mice completely derived from ES cells, they require specialized skills and/or equipment, or specific mutant mouse lines to produce blastocyst-stage embryos. This complexity may be unnecessary if ES cells with strong germline potential are employed, and the process described here is used to speed cohort generation.
Coat color of the resulting chimeras typically is used as an easy but indirect marker of the percentage of germ cells derived from the ES cells. For example, if embryos from a white mouse are injected with ES cells from a black mouse, resulting in a black-and-white coat, a preponderance of black patches is viewed as indicative of a high uptake of ES cells. In turn, chimeras with the most black fur would be selected as breeders. Yet, there is not always a strong correlation between coat color and the percentage of germ cells with the modified allele, which may take multiple rounds of breeding to discover.
Molecular analysis of sperm (genotyping) can help avoid this potential delay by ascertaining directly the ES cell contribution to the germline, providing a more accurate method for selecting male chimeras for breeding. Sperm is harvested from all sexually mature chimeras and cryopreserved using standard protocols. Genomic DNA is extracted from a sperm aliquot and quantitative PCR is used to assess the percentage of cells carrying the wild type and modified gene. Rather than rely on coat color as an indirect measure, this analytical approach provides a direct measure of a chimera’s appropriateness for breeding to attain the desired genotype efficiently.
In parallel, it is very useful to analyze the fertilization potential of the cryopreserved sperm sample by using an aliquot in a test IVF. Knowing which male is the most fertile and which has the highest likelihood of transmitting the ES cell genome to the next generation facilitates well-informed choices for cohort production. If the results indicate the clone(s) used in the blastocyst injection have poor germline potential and/or result in infertile or sub-fertile males, one can quickly select additional clones for chimera generation. If male chimeras with good germline potential and fertility are identified, one can move forward with greater confidence that the cohort will meet the study needs.
Speeding Cohort Delivery
After chimeras have been chosen for breeding, IVF can speed production of a study-size cohort considerably. The selected male’s sperm can be used to simultaneously fertilize hundreds of oocytes, generating many offspring faster and more predictably than with natural mating. IVF has other advantages: It occurs on a more predictable timeline, making it easier to plan for studies, and several rounds of chimera breeding can be skipped, saving three to four months. IVF also offers the flexibility to produce, then breed, mice at the most appropriate health status for the study.
To maximize the speed advantages provided by the strategy outlined above, it is advantageous to produce male chimeras without a selectable marker cassette, avoiding the need to delete the selection marker later through additional breeding. This can be accomplished through in vivo deletion in the male germ line using self-excising selection cassettes, or use of CRISPR in ES cells to eliminate the need for a selectable marker. Alternatively, in vitro deletion of a standard selection cassette in ES cells by recombinase transfection can be employed, but at the expense of some added time and effort. Lastly, if these strategies are not feasible, one could use oocytes derived from a recombinase-expressing mouse line for the IVF. However, in this case the deleter transgene or knock-in allele would need to be selected against during breeding.
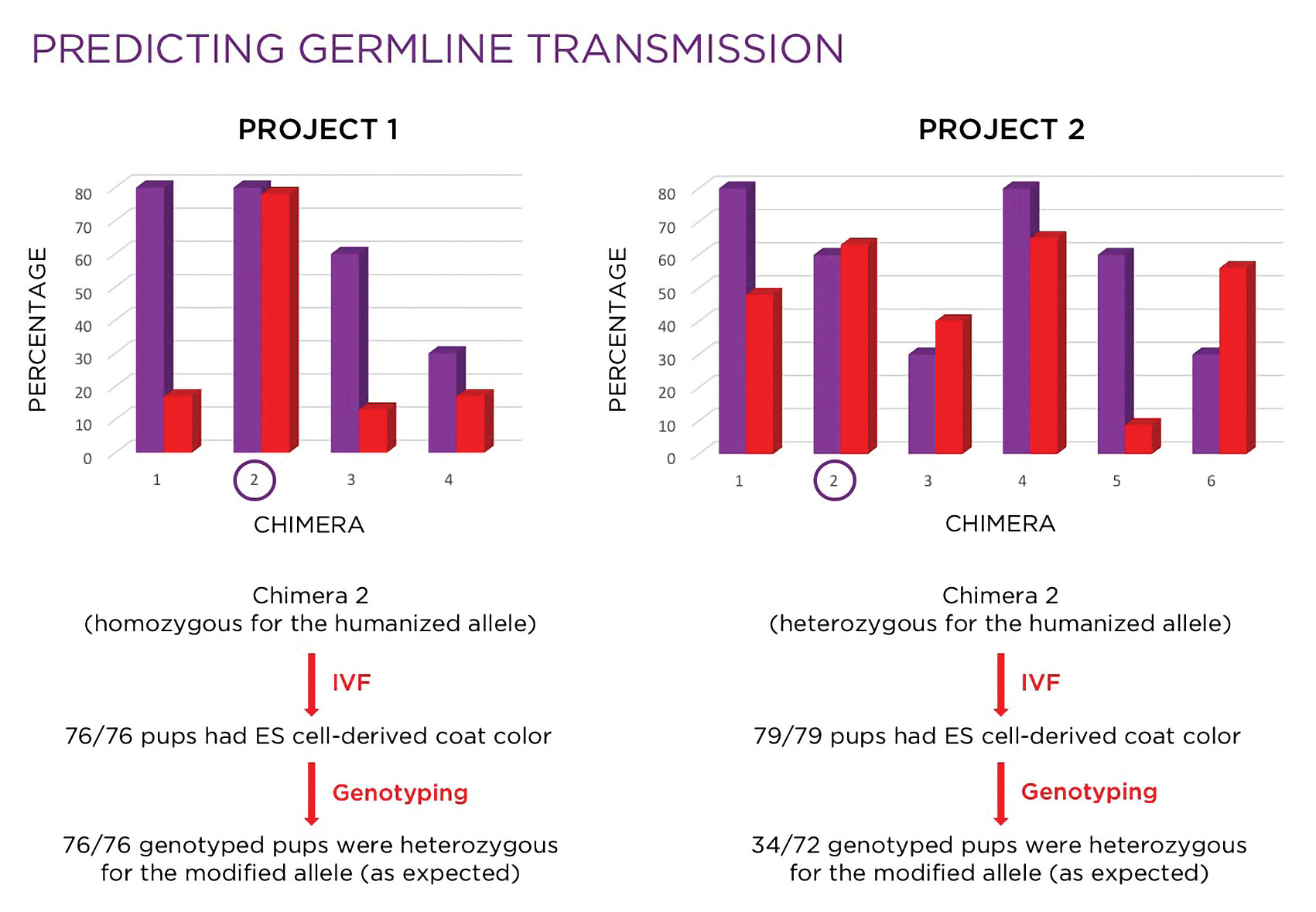
Figure 2. Investigators use molecular analysis to select the mice that are the most suitable candidates for breeding.
Results in Action
Data from a real-world custom model generation project demonstrates the advantages of the strategies outlined above.
As illustrated in Figure 2, based on coat color alone, chimeras #1 and/or #4 might have been selected for breeding, but molecular analysis revealed chimera #4 would be the better choice of the two. While chimeras #3 and #6 would likely not have been considered for breeding, molecular analysis indicated that both would be reasonable choices. In this case, chimera #2 was selected, based on the combination of a high fertility rate and high percentage of the modified allele in the germ line, and used in an IVF with oocytes derived from wild-type females. All 79 pups born were black, indicating 100% germline transmission of the ES cells. Of the 72 mice genotyped, 34 (47%) were heterozygous, which was consistent with expectations.
Sperm genotyping for selecting breeders, combined with IVF for rapid cohort expansion, is a rational approach to mouse model generation and breeding for complex genome modification projects requiring an ES cell-based targeting approach (Figure 3). This strategy can assist investigators in advancing preclinical studies sooner, making data-driven strategic decisions, and shortening drug discovery timelines.

Figure 3. To get the most productive mouse chimeras, a combination of sperm chimerism and in vitro fertilization is necessary.
Kenneth Albrecht, Ph.D. ([email protected]), is scientific program manager at Taconic Biosciences.



