February 1, 2010 (Vol. 30, No. 3)
Throughput, Resolution, and Multiplexing Drive Cost-, Time-, and Resource-Efficient Advances
The scope of real-time quantitative (qPCR) assays for gene-expression analysis in research applications and for sequence-specific nucleic acid detection for diagnostic purposes continues to grow. Driving this growth are increasingly robust, automated, and high-throughput technologies and multiplexed analytical strategies.
At Intelligent Enterprise Solutions’ “qPCR Symposium” held recently in Millbrae, CA, presenters explored advances providing improved qPCR throughput and quantitative resolution, discussed the challenges in preanalytical sample processing and the selection of a qPCR assay with high specificity, and described emerging application areas such as microRNA (miRNA) profiling in biological samples.
Melissa Kelley, Ph.D., R&D research scientist at Thermo Fisher Scientific, discussed the challenges in distinguishing between closely related members of a gene family when using qPCR in an interference RNA (RNAi) study to assess the phenotypic effects of gene knockdown.
As an example, she reviewed the use of Thermo Scientific’s Solaris qPCR gene-expression assays to study the regulation of a FOXO transcription factor (FOXO1), which is regulated via phosphorylation by AKT enzymes, a family of serine/threonine protein kinases. The AKT family comprises at least three highly homologous members that share 70–80% sequence identity. Silencing of AKT activity results in FOXO dephosphorylation, translocation of the transcription factor to the nucleus, and up-regulated gene expression. AKT dysfunction has been associated with various disease states, including cancer and diabetes.
In the experiment described by Dr. Kelley, small interfering RNAs (siRNA) were designed to knockdown the expression of each individual AKT family member. qPCR was then used to measure target gene knockdown, followed by high-content analysis to assess the biological phenotype. Dr. Kelley focused on the question of how to select a qPCR assay when the gene of interest has multiple splice variants, as do two members of the AKT family.
Solaris qPCR assays include probe/primer sets designed using a tier-based algorithm. Each assay detects all known splice variants of a gene and allows the researcher to discriminate between closely related members of a gene family. The assays are designed to target a consensus sequence created from a region common to all of the transcript splice variants. The assay kit provides the researcher with the probe and primer sequences to facilitate compliance with evolving MIQE guidelines.
To increase the sequence design space and to raise the melting temperature of bound complexes to allow for use of shorter, more specific probes, the design algorithm incorporates minor groove binder and Superbase technologies from Epoch Biosciences, now part of Nanogen. This strategy allows for greater than 97% coverage of both the human and mouse genomes. It also “minimizes amplification of contaminating genomic DNA and eliminates off targeting,” said Dr. Kelley. She presented relative gene-expression data generated using the Solaris qPCR assay demonstrating specific siRNA-mediated knockdown of a targeted AKT enzyme and expression levels of the other two AKT family members comparable to those measured in an untreated, control sample.
Analysis of the relative expression of AKT gene family members following RNAi-mediated knockdown, combined with biological assays to evaluate the cellular localization of the FOXO1 transcription factor, enabled the determination of AKT isoform-specific regulation of FOXO1 and illustrated the phenotypic effects of inhibiting the AKT pathway.
The experiment showed that silencing of individual AKT enzymes did not result in a significant change in the localization of FOXO1, supporting the theory that there is redundancy built into the AKT signaling pathway. In contrast, knockdown of combinations of two AKT family members, and simultaneous downregulation of all three enzymes led to increasing FOXO1 translocation from the cytoplasm to the nucleus.
Resolution and Throughput
Whether qPCR can achieve better than twofold discrimination and whether greater quantitative resolution is feasible in a cost-, time-, and resource-efficient manner were the questions put forth by Ken Livak, senior scientific fellow at Fluidigm. He described a study using qPCR-based copy number variation (CNV) determination as a paradigm for analyzing the parameters of quantitative resolution. The study was designed to determine how many experimental replicates would be needed to be able to distinguish between one, two, three, four, or five copies of an X-linked gene. The ability to differentiate four copies from five copies would correspond to a 25% difference, or 1.25 discrimination.
Livak performed the study using Fluidigm’s 96.96 Dynamic Array Integrated Fluidic Circuit (IFC), a microfluidic-based biochip that utilizes a matrix design to carry out 9,216 real-time qPCR experiments in parallel. The 96.96 Dynamic Array contains inlets for up to 96 different samples and 96 different assays, which are combined pair-wise.
In the CNV experiment, Livak evaluated five different samples—each contained one, two, three, four, or five copies of the X chromosome—and each sample was pipetted into 19 sample inlets. Each qPCR assay mixture was pipetted into 24 assay inlets, resulting in 19 x 24 or 456 replicates for each sample. A diploid gene—present in two copies/cell—was used as a reference calibrator.
Fluidigm’s other qPCR platform, the Digital Array IFC, yields a digital output and can perform endpoint PCR. A sample/reagent mixture introduced onto a chip is divided into hundreds or thousands of different chambers, with an identical volume in each chamber. Following PCR, a positive result will indicate a chamber containing one or more copies of the gene of interest, whereas a negative chamber will have zero copies of the gene. The system determines the number of target molecules per microliter of sample by counting the number of positive chambers, applying a Poisson distribution correction, and dividing the resulting number by the total input volume.
The company recently introduced the 48.770 Digital Array IFC, which can partition up to 48 different sample/reagent mixtures into 770 chambers per reaction. Livak described a CNV experiment performed using endpoint qPCR, in which a sample containing four copies of a target gene was compared to a sample with five copies of the target gene; the samples also contained a reference gene. Each sample was loaded into 24 panels of a 48.770 Digital Array and dispersed into 770 chambers.
The results demonstrated that 1.25-fold discrimination could be achieved using real-time PCR on the Dynamic Array with 18 replicates, while 86 replicates could distinguish 1.1-fold differences. In comparison, with digital PCR, a 1.25-fold difference could be discriminated using approximately 1,200 chambers, with 8,000 chambers needed to achieve a 1.1-fold discrimination.
Whereas, real-time qPCR offers advantages in throughput, digital PCR is easier to perform, with less chance for error, and it can be run as an endpoint assay, concluded Livak. “Quantitative resolution of qPCR can be enhanced to 1.25-fold, or even 1.1-fold, by running a large number of reactions,” which is feasible and practical on a high-throughput microfluidic platform.
Fluidigm is exploring the use of the Digital Array IFC for noninvasive prenatal analysis of fetal DNA. The goal is to develop a qPCR system that can reliably and cost-effectively amplify the small amounts of fetal DNA present in a mother’s blood, accurately quantify gene copy number, and distinguish, for example, the three copies of chromosome 21 that are present in a sample from a fetus with trisomy 21 (Down syndrome) from the two copies of chromosome 21 in the cells of a fetus without the genetic disorder.
High throughput real-time PCR is moving in two main directions, noted Matthias Hinzpeter, Ph.D., head of program management qPCR and NAP systems at Roche Applied Science: a high-density microplate-based array format or a microfluidics-based chip format. Roche’s newest LightCycler® PCR system, the LightCycler 1536, is a microplate-based qPCR platform that processes 1,536 reactions in less than 50 minutes. Reaction volumes range from 0.5 to 2.0 microliters.
“The major advantage of this system is the high flexibility with regard to number of targets and number of samples,” said Dr. Hinzpeter. “The technology breakthrough was the development of a 1,536-well microplate with rapid and homogeneous temperature distribution across the 1,536 wells with small reaction volumes.”
Roche’s Universal ProbeLibrary qPCR assays are based on 165 prevalidated probes suitable for studies of samples from a variety of organisms. The probes incorporate locked nucleic acid (LNA) technology for increased specificity and sensitivity and to allow for higher melting temperatures. Universal ProbeLibrary Reference Gene Assays allow for multiplex assays to quantify expression levels of human, mouse, or rat genes.
Probing Clinical Samples
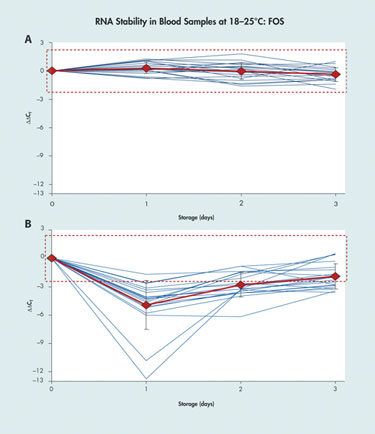
The use of real-time qPCR to detect and quantify RNA sequences in blood, tissue, or other types of biological samples is complicated by the instability of the RNA and the changes in transcript concentrations that can occur between sample collection and testing due to post-collection gene induction, cell death, and enzymatic degradation.
“A change in the CT [cycle threshold] value of 10–12 in the sample material is not unusual,” said Ralf Wyrich, Ph.D., associate director R&D, diagnostic sample preparation and stabilization at Qiagen. The CT is a representation of fluorescent signal accumulation, with CT values being inversely proportional to the amount of target nucleic acid in a sample.
Dr. Wyrich described a technique for post-collection stabilization using the PreAnalytiX PAXgene blood RNA tube, which preserves the transcript profile of cellular RNA in blood for up to three days at 18–25ºC, for up to five days at 2–8ºC, and for long-term archiving at -20ºC to -70ºC. The PAXgene blood-collection and RNA-stabilization system has received U.S. FDA clearance and CE mark in Europe for use in molecular diagnostic testing based on real-time PCR.
He also presented data demonstrating an approximate 1 log enrichment of miRNA on average from blood collected in a PAXgene Blood RNA tube compared to standard RNA-extraction procedures using a dedicated sample-preparation kit for the isolation of small RNA species.
A second PAXgene product was designed to preserve the RNA in bone- marrow samples, and the newest PAXgene system targets the collection and preservation of tissue samples to allow for both classical histomorphology and PCR-based molecular analysis to be performed on the same specimen.
The commonly used method for tissue preservation relies on formalin fixation, which does not stabilize the RNA content of the sample and, furthermore, preserves the tissue through chemical crosslinking of molecules, which interferes with qPCR analysis. Currently available nucleic acid stabilization technologies for use in tissue samples do not preserve the morphology of the tissue material.
The PAXgene tissue container exposes the sample first to a fixation reagent and then to a stabilization reagent. In this way the same sample can be sectioned for histomorphologic analysis and processed with either a RNA, DNA, or miRNA kit for nucleic acid purification and analysis.
“The lack of standardization in preanalytical sample handling was recognized by the European Commission, which funds several projects in this field,” said Dr. Wyrich. Qiagen is spearheading a European consortium called SPIDIA (standardization and improvement of generic preanalytical tools and procedures for in vitro diagnostics), which is funded by the European Union, with a total four-year budget of €13 million ($18.7 million). The goal of the 16 participating companies and institutions representing 11 countries is to standardize preanalytical procedures and protocols for molecular diagnostic applications.
Comparison of Qiagen’s PAXgene blood RNA tube (A) with standard EDTA blood collection tube (B)
Quantifying miRNA
qPCR is a potent technique for microRNA expression profiling to assess gene regulation, yet the characteristics of miRNAs present challenges for assay design. miRNAs are only 19–22 nucleotides in length, making design of DNA primers without overlap a challenge.
In addition, they have high sequence homology, and miRNAs within a family may differ by only a single nucleotide. They also have a wide range of GC content, from as little as 25% to as much as 98%. Low GC content makes it difficult to design primers with a melting temperature high enough for PCR, whereas a high GC content makes it difficult to avoid background interference from the formation of primer dimers.
By introducing locked nucleic acid (LNA™) bases into both the forward and reverse PCR primers included in its new miRCURY LNA™ Universal RT microRNA PCR system, Exiqon was able to design short primers with high specificity and sensitivity for miRNA analysis.
“LNAs increase sensitivity and help discriminate between mismatches,” said Ditte Andreasen, Ph.D., senior scientist at Exiqon. She presented data showing that including LNAs in PCR primers increases the sensitivity by about 100 times in comparison to using DNA primers. She also showed how LNAs can lead to greater discriminatory power between single nucleotide mismatches by increasing melting temperature between a mismatch and perfect match target.
Exiqon’s new miRNA qPCR platform incorporates the one-tube Universal RT (reverse transcription) reaction for cDNA synthesis, eliminating the need to perform multiple gene-specific reverse transcriptase reactions and preamplification, according to the company. A single Universal RT reaction performed on an RNA sample as small as 40 ng generates enough material for two 384-well plates, allowing for qPCR analysis of 730 different miRNAs.
Dr. Andreasen reported the results of studies using the LNA Universal RT system to profile miRNA expression in clinical samples: formalin-fixed paraffin-embedded tissue samples, as well as blood serum and plasma samples. She demonstrated the ability to quantify miRNA expression levels compared to reference standards, to discriminate single nucleotide differences, and to generate miRNA signatures. She described efforts under way at the company to develop colon cancer diagnostic qPCR assays using the system, assessing disease-related miRNA expression in biopsy samples.
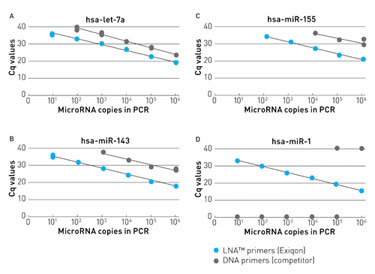
Using LNA™-enhanced primers for microRNA qPCR results in greatly increased sensitivity compared to DNA primers, according to Exiqon.
Analyzing Proteins with PCR
TaqMan® Protein Expression Assays from Applied Biosystems, part of Life Technologies, “open up new territory for real-time PCR,” facilitating quantification and analysis of protein expression as well as analysis of protein-protein interactions and pathway responses resulting from post-translational phosphorylation events, contends David Ruff, Ph.D., principal scientist at Life Technologies.
At the heart of this homogeneous assay system are two antibodies, one of which contains a bound 3´ open end of an oligonucleotide and the other the 5´ open end of a different oligonucleotide. Each antibody recognizes a distinct epitope on the protein of interest. When both antibodies are bound to the target, their conjugated oligos are brought into close proximity; ligation of the oligos is then directed with a connector oligo sequence creating an amplicon template for primer and probe hybridization for real-time PCR amplification.
The antibody pair sets can be monoclonal or polyclonal antibodies; each antibody is labeled with biotin. They are conjugated to streptavidin-linked oligonucleotides. The entire workflow, encompassing preparation of the whole cell lysate, binding of the oligo-antibody probes to the target proteins in the wells of a microplate, ligation and inactivation of ligase, and PCR takes about 3.5 hours, according to Dr. Ruff.
The assay “requires very little material, about three orders of magnitude less than for a Western blot.” Whereas the limit of detection of a Western blot is typically 30,000–10,000 cells/lane, it is typically 10–35 cells/well for the protein assay. Using the resulting CT values, the research “can correlate protein to mRNA levels on one platform.”
Dr. Ruff presented an example of how the assay could be used to detect and quantify changes in expression of protein markers of stem cell pluripotency and differentiation. Another application he described used the Protein Expression Assay to measure changes following siRNA knockdown of a target mRNA. He provided data showing a good correlation between the results of an in situ hybridization experiment and the corresponding protein expression assay results.
Life Technologies’ current product line includes six assays for stem cell regulatory proteins. In development and expected to launch in mid-2010 is the TaqMan Protein Expression Assay Open Kit, for which customers will supply the biotin-labeled antibodies for their protein target of interest, and the company will provide the streptavidin-labeled oligos and Universal assay reagents for ligation and qPCR.
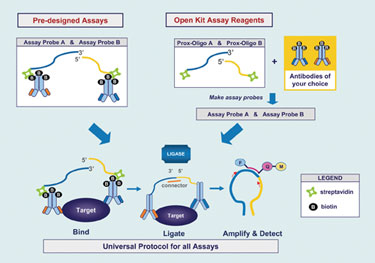
TaqMan® protein-expression assay products from Applied Biosystems include six predesigned assays that measure relative protein expression.



