December 1, 2014 (Vol. 34, No. 21)
Back to Basics: Using a Fixed-Path-Length Spectrophotometer in the UV Range
With the advent of new technologies that aim to more clearly discern subtle and complex information from small samples, the requirement for setting quality control processes has never been greater. The need to determine sample concentration has particularly expanded in proteomics and genomics with both areas requiring lengthy sample preparation and data analysis. Set as an intermediary step, protein or nucleic acid quantitation plays a key role in the outcome of many downstream experiments.
The way in which the quantitation is performed is equally important: the quality control process that can potentially quantify and qualify vast amounts of sample types should be quick and simple.
Biochrom’s BioDrop μLite is a split-beam UV-Vis spectrophotometer equipped with a built-in microvolume sample port, specifically designed for the quantitation of small sample volumes such as proteins or nucleic acids. Set to work with as little as 0.5 μL of sample, BioDrop μLite delivers quick and high-quality measurements by setting only a single and fixed path length so as to reduce bias and inaccuracy related to path length design.
We present here how the BioDrop microvolume sample port can provide valuable information for the quantitation of DNA samples at concentrations ranging from a few nanograms per microliters up to 2,500 ng/μL of dsDNA at 260 nm, achieving minimal sample carryover and high precision, well within the measurement capabilities of the instrument.
Material and Method
Sample preparation
Lyophilized pure genomic DNA sample (dsDNA) from calf thymus was weighed with a precision scale (d=0.1 mg) and diluted to 1,000 mg/mL in Tris-EDTA buffer solution pH 7.4.
The master sample was subsequently diluted for constituting new samples. Each sample was vortexed and centrifuged.
The samples were measured using 1.5 μL of solution each time. The microvolume sample port was wiped using lint-free tissue after each sample measurement.
Result and Discussion
BioDrop detection range, where purity ratios matter
According to the Beer-Lambert law, measuring absorbance values allows for the determination of sample concentration at a given wavelength. This law applies to direct measurements of nucleic acids where the peak absorbance at 260 nm is used to calculate the sample concentration. Assuming the sample is DNA, the sample concentration can be defined by using the multiplication factor 50, divided by the path length of the BioDrop microvolume sample port: 0.05 cm.
Linear precision can be reached within the measurement range of the microvolume sample port of the BioDrop spectrophotometer (Figure 1).
As absorbance at 260 cannot always be specific to DNA, a more comprehensive way to analyze nucleic acid samples is also to calculate the purity ratio at 260/280 and 260/230 reported in the Table.
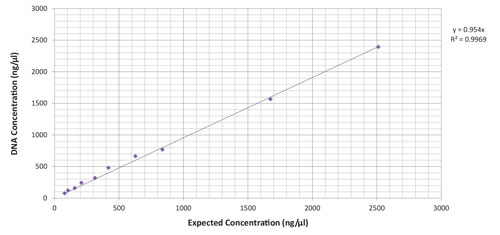
Figure 1. BioDrop microvolume sample port linear detection range. dsDNA samples were obtained via dilution series of master sample, which consisted of precisely measured 0.105 g of lyophilized dsDNA sample diluted in Tris-EDTA buffer ph 7.4 so as to reach a total volume of 10 mL. Sample measurements were taken at 260 nm.
The first ratio concerns protein contamination in DNA samples. Pure DNA samples have a ratio 260/280 greater than 1.8. The samples can be equally qualified as free from proteins if the values reach 2.0 as a maximum. When protein residues are present, the 260/280 ratio is shifted as an indication of the contamination.
The second ratio 260/230, concerns organic contaminant and solvents detection such as phenol, thiocyanates, urea, carbohydrates, B-mercaptoethanol, and guanidinium residues from chaotropic lysis buffer, which are often responsible for poor DNA detection in further DNA sequencing processes by significantly affecting the PCR reactions.
If those ratios are checked against the sample concentration, within the detection range of BioDrop (Table), a rapid estimation of the sample quality can be made. BioDrop can accurately provide purity ratio within a 1,000 dynamic range for DNA samples having concentrations ranging from 2.0 ng/μL up to 2.4 μg/μL.
Purity ratios are critical to insuring both accurate quantitation and identifying contaminants that can significantly interfere with downstream experiments such as those involving PCR, protein function assays, and transfections, for example. BioDrop systematically provides sample purity ratios when using its preprogrammed nucleic acid quantitation methods such as those for single-stranded or double-stranded DNA, RNA, or oligonucleotides. Those ratios are recorded for every sample each time a measurement is taken.
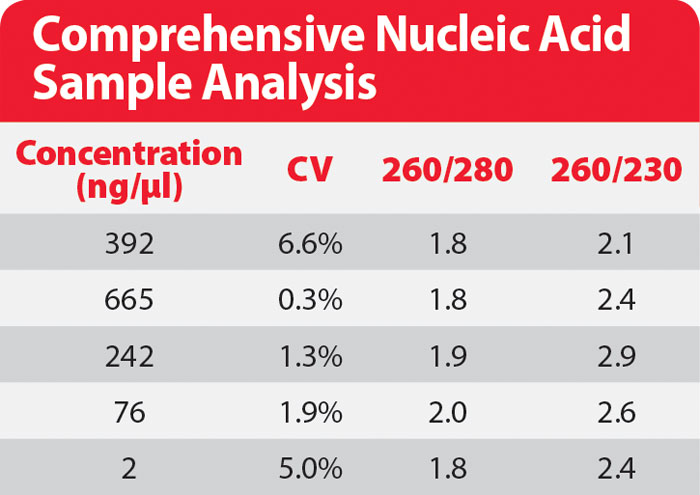
dsDNA concentrations and purity ratios of uncontaminated samples obtained from BioDrop microvolume sample port. Pure dsDNA samples diluted in Tris-EDTA buffer ph 7.4 were measured using the BioDrop microvolume sample port where absorbance measurements at 260, 280, and 230 nm were systematically taken. The purity ratios (A260/A280 and A260/A230) were directly calculated by the BioDrop Resolution software.
BioDrop Accuracy and Measurement Precision Is Designed into the Instrument
The BioDrop microvolume sample port allows the light beam to cross the sample horizontally. The crossing distance is static and is the space in between the two unmovable measuring probes. It also represents the instrument fixed 0.05 cm path length. If the volume of sample deposited on the microvolume sample port is sufficient to reach the measuring probes, sample measurement can be performed with minimum variability. If homogenous samples are measured repeatedly, the precision of the measurement of sample concentration is optimized (Table and Figure 2). With this specific design, the effect of the path length reduction is minimized so that it does not account for the overall variability observed within a particular sample set. Also, the design of the microvolume sample port is optimized for minimum sample carryover (Figure 2).
For sample with reduced viscosity, a minimum of 0.5 μL of solution should be enough, or 1.5 μL can be used.
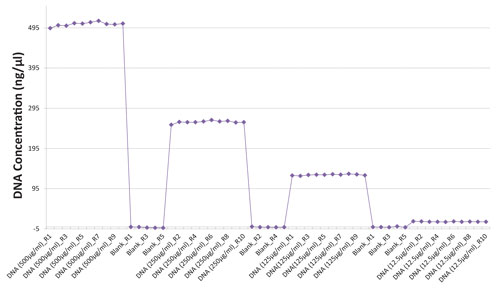
Figure 2. BioDrop microvolume sample port sample carryover test. dsDNA samples were obtained via dilution series of master sample, which consisted of precisely measured 0.105 g of lyophilized dsDNA sample diluted in Tris-EDTA buffer ph 7.4 so as to reach a total volume of 10 mL. Sample measurements were taken at 260 nm.
Conclusion
The BioDrop microvolume sample port measures accurate absorbance values of dsDNA samples starting from a few nanograms per microliters up to 2,500 ng/μL, covering more than a 1,000-fold linear dynamic range.
As DNA absorbance measurement in the UV range is not as straightforward as other alternative absolute quantitation methods, visualization of complementary information such as purity ratios is equally important, making this absorbance measurement method indispensable for assessing sample quality. Added to this, purity ratios cannot be obtained via absolute quantitation methods and BioDrop offers advantage for performing quality control of small volume samples by providing valuable sample information over a very large concentration range.
By designing the BioDrop Platform with a single and unmovable path length, accurate data comparison is afforded, which is especially appropriate for quality control requirements of downstream experiments. As precise and accurate absorbance data is not only very valuable information for dsDNA samples, BioDrop improved fixed path length also has pre-recorded quantitation methods to measure microliter-sized samples of ssDNA, RNA, oligonucleotides, and proteins, allowing BioDrop to remain accurate over time, eliminating the need for sample port recalibration.
Fabiola Saffache ([email protected]) is associate product manager molecular analysis, Biochrom.



