March 1, 2007 (Vol. 27, No. 5)
Steve Harrison
Graham Lidgard Ph.D.
Pathogen Sequence Variability Requires Flexibility in Assay Design
Real-time PCR has gained significant acceptance by high-complexity clinical laboratories developing tests for a variety of applications. Sensitivity, automation, and turnaround time have made PCR particularly useful for pathogen detection, especially for organisms difficult to grow in culture.
However, strain variability of genetic sequences from pathogens complicates probe and primer design and interpretation of results. While designers typically target regions that are conserved across various strains, these regions can be too short or problematic for traditional real-time PCR chemistries. Additionally, variants with unknown polymorphisms in these conserved regions may escape detection.
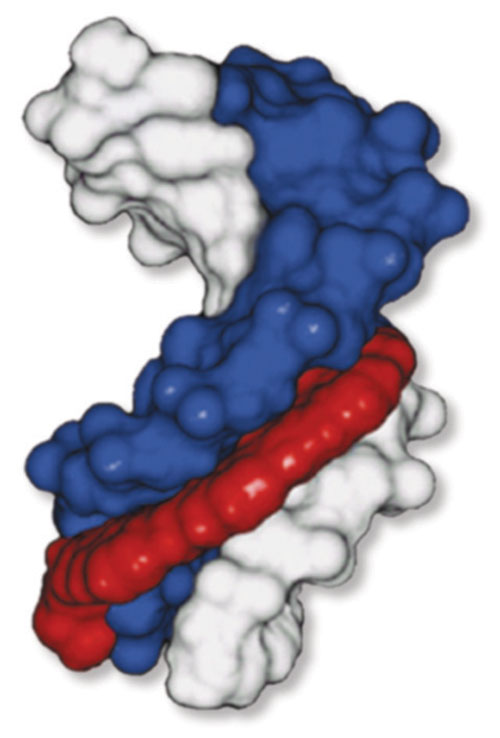
Figure 1
Dealing with Problematic Regions
Nanogen (www.nanogen.com) developed new technologies to overcome these obstacles. MGB™ Technology utilizes a minor groove binder attached to single-stranded DNA probes to boost the stability and raise the melting temperature (Tm) of the DNA duplex formed when probes bind to complementary targets. This enables the use of smaller probes capable of detecting short conserved regions when developing assays to detect multiple strains (Figure 1).
Nanogen Superbases—Super A and Super T—fine-tune the Tm of probes or raise the binding stability of primers. When incorporated into synthetic oligonucleotides, these modified nucleotides display increased binding affinity for complementary nucleotides, providing additional duplex stability. As a result, Superbases offer further flexibility when designing primers or probes to short, conserved regions or regions rich in adenosine or thymidine residues (Figure 2).
Other examples of problematic regions that reduce probe and primer design flexibility—and ultimately the availability of high-performance assays—include those rich in cytosine residues and those that include polymorphic sites. C-rich regions require guanidine-rich probes and primers, which can be difficult to manufacture and can prevent efficient hybridization. Substituting Super G for guanosine in probes and primers overcomes both the manufacturing and hybridization challenges. Additionally, Super N is a universal base that binds efficiently to all four bases, eliminating the negative impact of known polymorphic sites.
Finally, even when all of these complicating design factors are addressed, there still remains the possibility of a variant sample containing an unknown polymorphism. When this occurs within the region targeted by the probe, results can be difficult to interpret or, even worse, falsely called as negative. Attaching the MGB® to the 5´-end probes, however, as is the case with MGB Alert® Detection Reagents (Figure 3), prevents probe degradation by the 5´ exonuclease activity of Taq polymerase, leaving the probe intact for post-PCR melt curve analysis. Melt curve analysis provides additional test information and greater confidence in results without additional expense or technician time.
Enterovirus Amplification
A good example of this is demonstrated by work performed during the development of probes and primers designed to amplify and detect a broad range of enterovirus types and their related strains. A 16-bp probe was designed to target the highly conserved 5´ untranslated region of the enterovirus genome using publicly available consensus sequence data. However, during the course of running numerous clinical samples, several variants that compromised sensitivity and interpretation of data were identified through the use of melt curve analysis. Sequencing of the strains confirmed the results obtained using melt curve analysis.
Recognizing that labs would prefer to avoid dealing with these challenging design issues, Nanogen has incorporated these technologies into the MGB Alert® Detection Reagents. These analyte-specific reagents consist of probes and primers designed to allow labs to develop protocols to amplify and detect genetic sequences associated with many of the commonly tested for viruses and bacteria such as enterovirus, CMV, HSV, Bordetella pertussis, and many others. They are compatible with most commercially available instrumentation and come in ready-to-use formulations.
With these reagents, research and clinical labs can develop higher performance tests that address the variable nature of pathogens and take greater advantage of the power offered by real-time PCR.
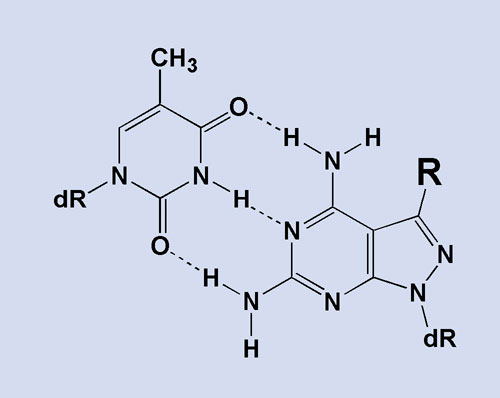
Figure 2
Figure 3
Steve Harrison is director of marketing, real-time PCR products and Graham Lidgard, Ph.D., is senior vice president of research & development at Nanogen.
E-mail: [email protected].
Web: www.nanogen.com.