June 15, 2013 (Vol. 33, No. 12)
Laura Whitman DNA2.0
Medini Gore DNA2.0
Jon Ness, Ph.D. DNA2.0
Elias Theotdorou, Ph.D. DNA2.0
Claes Gustafsson Ph.D. COO and Founder ATUM
Jeremy Minshull, Ph.D. DNA2.0
While the traditional cloning workflow has been used in laboratories for decades, new techniques have emerged that allow for rapid assembly of multiple genetic fragments. The Electra Vector System, developed by DNA2.0, is a universal cloning system that utilizes the type IIS restriction enzyme SapI in a single-tube reaction (Figure 1). In this reaction, SapI recognizes a 7bp nonpalindromic recognition sequence in the cloning vector, and leaves a 3 bp 5´ overhang after digestion for ordered assembly. This process leaves no cloning scars and does not require PCR or other mutation-inducing amplification.
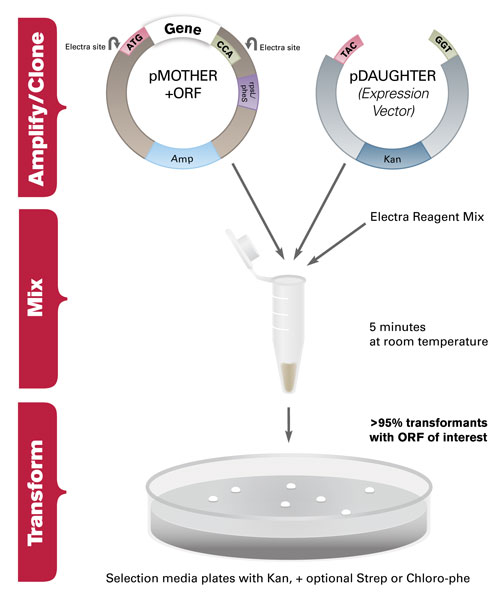
Figure 1. Single tube digestion and ligation reaction using the Electra System: The MOTHER vector, carrying the gene of interest, is mixed with linearized DAUGHTER vector in the presence of Electra Reagent Mix. The reaction is incubated for 5 minutes at room temperature, transformed into E. coli and plated on LB/agar plates with antibiotic.
A collection of IP-Free© bacterial, mammalian, and yeast DAUGHTER expression vectors have been constructed to function as Electra vectors. Furthermore, any vector can be easily converted by DNA2.0 to function as an Electra vector. A gene that is provided in a MOTHER vector can be quickly and efficiently moved into any DAUGHTER vector (Figure 2), allowing the gene to be tested under different conditions, including promoters, ribosome binding sites, C- and N-terminal tags, and/or fusions. In this experiment, the gene encoding KringleYFP (yellow fluorescent protein) was cloned into a series of DAUGHTER vectors and then analyzed.
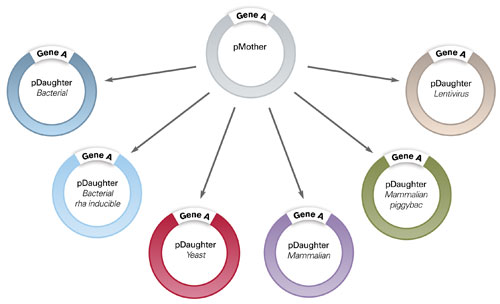
Figure 2. Efficient transfer of your ORF into several host expression systems using the Electra System. An ORF of interest (Gene A) can be easily transferred from the MOTHER vector (pMOTHER) into a range of DAUGHTER vectors (pDAUGHTERs). DAUGHTER vectors offer a variety of resistance markers, promoters and C- and N-terminal tags and/or fusions.
Results
A MOTHER vector carrying a gene encoding KringleYFP was mixed with an E. coli expression DAUGHTER vector in the presence of Electra Reagent Mix for 5–60 minutes. Reactions were transformed into E. coli and plated onto LB plates supplemented with appropriate selective antibiotics (Figure 1).
Transformants resulted from reactions that were incubated for as little as 5 minutes and maximum efficiency was obtained by 40 minutes (Table 1). Almost all transformants showed an inducible fluorescent yellow phenotype, indicating accurate movement of the gene from the MOTHER into the expression DAUGHTER vectors. DAUGHTER constructs are selected because of antibiotic-resistance markers.
MOTHER vectors carry a counter-selection marker, either rpsL (streptomycin sensitivity)1 or PheS (phenylalanine analog p-chlorophenylalanine sensitivity)2 that allows selection against carryover of MOTHER vector by plating transformants onto media that contains both DAUGHTER selection antibiotic and MOTHER counter-selection agent.
In a separate experiment, crude PCR product of the KringleYFP gene was successfully cloned into an E. coli DAUGHTER vector without any PCR reaction treatment or cleanup, using a single-tube 5 minute protocol.
Summary
The Electra Vector system enables quick and efficient transfer of genes into any DAUGHTER vector(s), either from a MOTHER vector or directly from a PCR reaction. This facilitates convenient testing of expression system parameters and enables rapid optimization of genetic constructs. Unlike alternative systems, there are no intellectual property entanglements, no unwanted mutations from error-prone polymerases, and no sequence scars to affect expression and function.
The Electra system can also be used to combine multiple sequence elements simultaneously, facilitating the easy construction of combinatorial libraries.
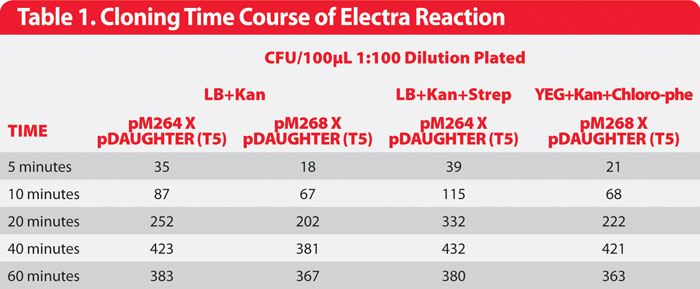
Table 1: Cloning Time Course of Electra Reaction
Experimental Protocol
Transfer of a KringleYFP ORF from MOTHER vectors to DAUGHTER vector:
- pMOTHER constructs: a yellow fluorescent protein gene (KringleYFP) was cloned into two MOTHER vectors:
- pMOTHER264 (pUC bacterial origin, Amp antibiotic resistance, rpsL counter-selection marker)
- pMOTHER268 (pUC bacterial origin, Amp antibiotic resistance, PheS counter-selection marker)
- SapI-linearized pDAUGHTER vector:
- pDAUGHTER441-SR (inducible T5 promoter, strong RBS, Kan resistance, pUC bacterial origin)
Reactions for the exchange of ORF from pMOTHER264 to pDAUGHTER441-SR (reaction 1) and pMOTHER268 to pDAUGHTER441-SR (reaction 2) were each set up in a 1.5 mL microfuge tube as follows (Table 2):
- Reactions were incubated for 5, 10, 20, 40 or 60 minutes at room temperature.
- 2.5 μL of each reaction was transformed into 50 μL of NEB 10-beta competent E. coli.
- 100 μL were plated onto LB agar plates with 30 μg/mL kanamycin or with 30 μg/mL kanamycin plus 100 μg/mL streptomycin. 100 μL reaction 2 were plated onto YEG agar plates, or YEG with 10 mM p-chlorophenylalanine.
- Incubated at 37°C overnight.
We also demonstrated that PCR product can be cloned into a DAUGHTER vector in 5 minutes at room temperature without any PCR reaction cleanup (data not shown).
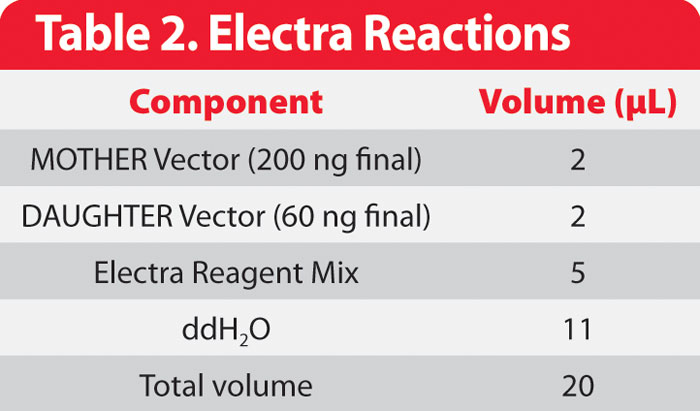
Table 2: Electra Reactions
DNA2.0
1140 O’Brien Drive, Suite A
Menlo Park, CA 94025
Claes Gustafsson, Ph.D.
COO
[email protected]
www.dna20.com/electra
References
1. Drecktrah et al, 2010. Appl Environ Microbiol 76(3): 985-987
2. Schneider et al, 1997. Gene 197: 337-341