August 1, 2017 (Vol. 37, No. 14)
Kristin Huwiler Global Strategic Manager Promega
James Vasta Ph.D. Research Scientist Promega
Cesear Corona Staff Scientist Promega
Chad Zimprich Research Scientist Promega
Jennifer Wilkinson Research Scientist Promega
Matthew Robers Senior Research Scientist and Group Leader Promega
NanoBRET Target Engagement Assays
Engagement of intracellular targets is often critical to drug mechanisms. Establishing a target engagement (TE) profile is an important step in small- molecule drug discovery as well as chemical probe development.1 TE involves monitoring the binding between protein targets and small molecules inside live cells. In drug discovery, TE plays a central role as it enables one to determine mode of action and then to correlate to functional efficacy.2
Ideal TE methods should possess the ability to 1) quantitate target occupancy or binding between the target protein and the small molecule in live cells, rather than lysates; 2) determine the selectivity of the small molecule for other closely related proteins in live cells; and 3) measure the direct or proximal binding between target and small molecule in live cells, rather than monitor a distal event that can be affected by multiple pathways.1
There are several methods to measure TE in live cells, including but not limited to affinity-based chemoproteomics, cellular thermal shift assays (CETSAs), and enzyme fragment complementation-based ligand stabilization.2 Most of these techniques fail to meet the qualities of an ideal TE assay as described above.
Live-Cell Quantitative Target Engagement Assay for Drug Discovery
Here we describe an ideal method, the NanoBRET™ Target Engagement Assay, which is a quantitative, direct-binding technology capable of integrating compound selectivity across closely related family members. The assay is applicable to many target classes and is scalable for multiwell formats.
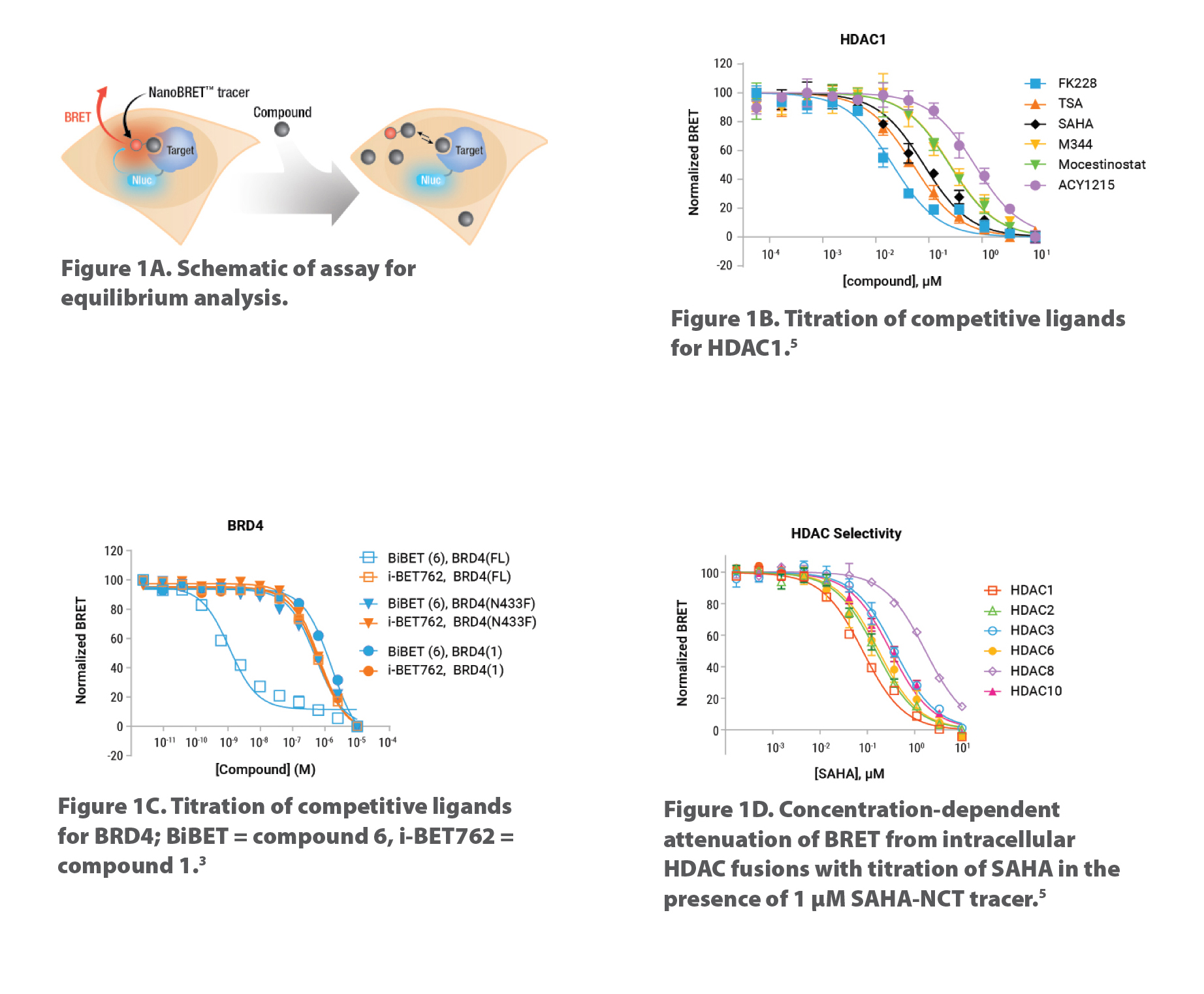
Using bioluminescence resonance energy transfer (BRET), we developed the first quantitative TE method capable of measuring compound binding to selected target proteins in live cells without disrupting the cell membrane. Four key components of the assay include: 1) cellular-expressed target protein fused to the very bright NanoLuc® luciferase; 2) cell-permeable fluorescent tracer that reversibly binds to target protein; 3) substrate for NanoLuc luciferase; and 4) Extracellular NanoLuc Inhibitor to ensure signal originates from live cells. Figure 1A shows a schematic of the competitive binding assay. Representative competitive binding data for HDAC1 and BRD4 expressed in live cells are shown in Figures 1B & 1C, respectively.
NanoBRET TE assays work with full-length, wild-type, truncated, or point mutant target proteins to better understand compound mechanism of action. Waring et al.3 used full-length BRD4 as well as truncated and point mutant variants to decipher a BiBET ligand that displays highest affinity when both domains of BRD4 are engaged simultaneously (Figure 1C).
We developed broad-coverage cell-permeable NanoBRET fluorescent tracers and NanoLuc-target fusion DNA constructs for assessing selectivity across closely related target proteins, such as the HDAC class I and IIb family members (Figure 1D) and BET Bromodomain family. These assays are commercially available. Newer NanoBRET TE Assays for new target classes are in development, which may, along with NanoLuc-target fusion DNA constructs, be sourced through Promega’s Custom Assay Services.
Figure 1. Schematic of NanoBRET TE equilibrium binding and representative competitive binding data for HDAC and BRD target classes. To determine quantitative target engagement via competition, compounds are added to cells with the fluorescent tracer. Any compound that is cell permeable and binds the target protein will compete with the tracer and result in a decrease in BRET signal. One day prior to compound addition, cells are transiently transfected with the target-NanoLuc fusion DNA construct. Representative BRET measurements of competitive binding data for HDAC1 (1B) and BRD4 (1C) expressed in live HELA and HEK293 cells, respectively.
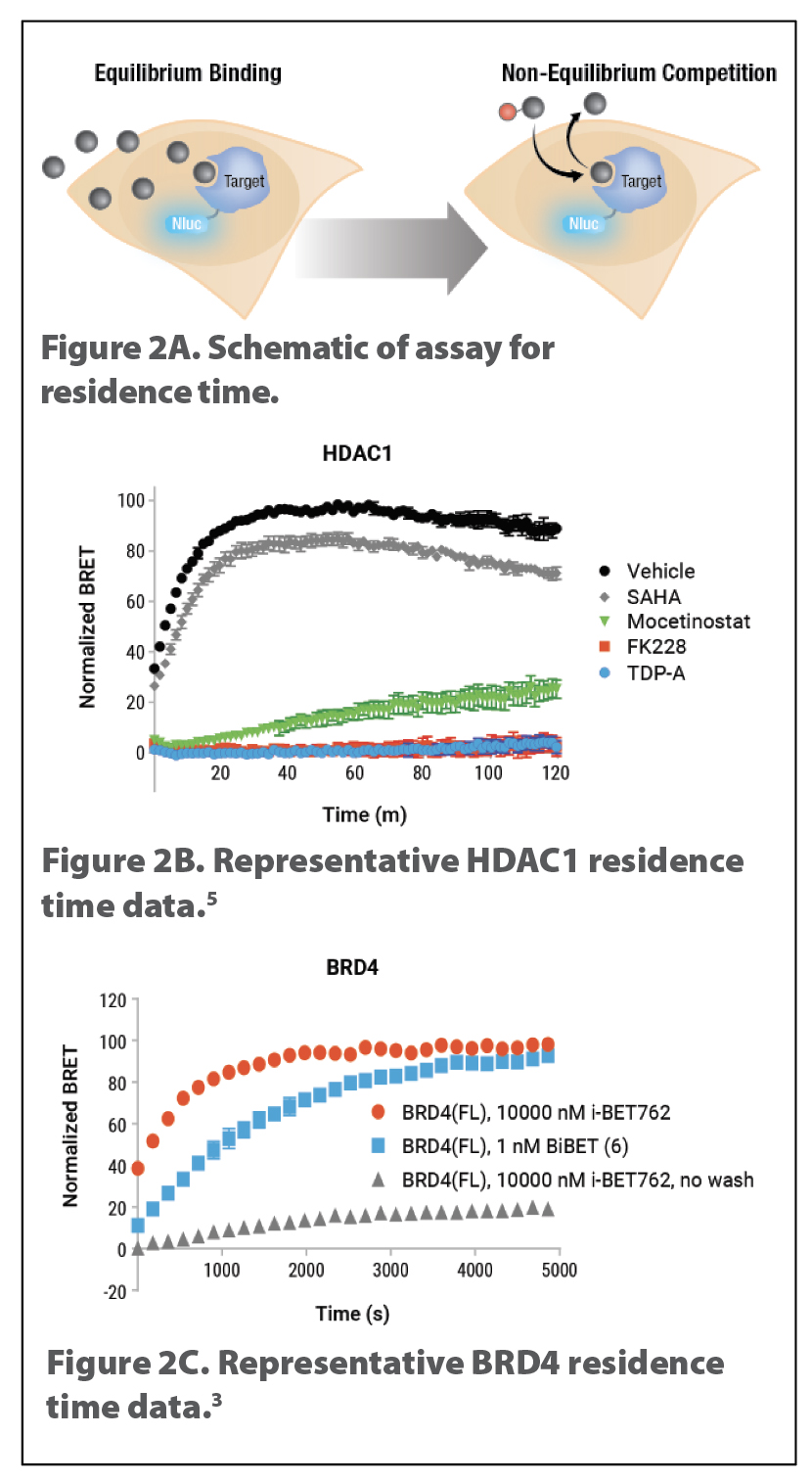
Residence-Time Analysis in Live Cells
Residence time is the lifetime that a ligand remains bound to its target protein under nonequilibrium, open conditions.4 Drugs are designed to be used in living systems where they are subject to nonequilibrium conditions associated with absorption, distribution, metabolism, excretion, etc. Therefore, residence-time analysis is an important parameter to modulate drug candidate efficacy and toxicity.
Fast binding of tracers and long-signal half-life of NanoLuc enable live-cell residence-time analysis (Figure 2A). The faster a compound dissociates from its target, the sooner the tracer can bind, resulting in an increase in BRET signal as a function of time. Representative NanoBRET residence-time analyses for several compounds against HDAC1 (Figure 2B) or BRD4 (Figure 2C) were published by Robers et al.5 and Waring et al.,3 respectively.
The technique may also be used to better understand mechanisms of action. For example, the approved drug FK228 (Romidepsin) shows prolonged effects at inhibiting HDAC activity in cells. We used NanoBRET to determine whether this action is due to long residence time.5 FK228 had the highest affinity of the compounds tested against HDAC1 (Figure 1B) as well as very long residence time (Figure 2B).
Figure 2. Schematic of NanoBRET TE residence time for HDAC and BRD target classes. Residence-time analysis using NanoBRET TE assay is performed by incubating test compounds with the cells expressing the target-NanoLuc fusion protein, followed by removal of excess com-pound, addition of tracer and substrate, and recording of BRET data kinetically (2A).
Summary
The NanoBRET TE method allows direct quantitative measurement of compound affinity for a target protein under physiological conditions, measurement of compound selectivity against closely related target protein family members, and a real-time assessment of compound residence time all in live cells. Because NanoLuc is so bright, target protein-NanoLuc fusions do not need to be expressed at high levels. Data shown here were conducted in 96-well assay plates, and the assays are scalable to 384-well or higher density, making it suitable for chemical probe development and multiple phases of drug discovery.
Kristin Huwiler ([email protected]) is Global Strategic Manager at Promega.
References
1. Simon GM, et al. Nat. Chem. Biol. 9:200 (2013), Determining Target Engagement in Living Systems.
2. Schurmann M, et al. Cell Chem. Biol. 23:435 (2016), Small-Molecule Target Engagement in Cells. DOI: 10.1016/j.chembiol.2016.03.008.
3. Waring MJ, et al. Nat. Chem. Biol. 12:1097 (2016), Potent and Selective Bivalent Inhibitors of BET Bromodomains. DOI: 10.1038/nchembio.2210.
4. Copeland RA. Nat. Rev. Drug Disc. (2016), The drug-target residence time model: a 10-year retrospective. DOI: 10.1038/nrd.2015.18.
5. Robers MB, et al. Nat. Comm. (2015) Target engagement and drug residence time can be observed in living cells with BRET. DOI: 10.1038/ncomms10091.







