January 15, 2012 (Vol. 32, No. 2)
Using Data Obtained from the Study of Metabolic Network Models
Novel recombinant protein therapeutics and the increasing demand for existing molecules continuously raise the need for higher biopharmaceutical manufacturing capacities.
This bolsters the need for efficient and robust bioprocesses. These are required for each individual molecule to ensure consistent product quality and to avoid interruptions in market supply due to production failures.
Consequently, biopharma companies and regulatory authorities like the FDA have increased efforts toward improving the characterization and the mechanistic understanding of manufacturing processes in recent years.
Widespread use of scale-down models and initiatives like PAT or QbD have resulted in growing collections of physicochemical product information and high-quality fermentation data. This often includes on-line signals as well as transcript and metabolite measurements.
Such data potentially represents a rich resource for increasing the speed and efficiency of process development. However, evaluation and interpretation of the acquired data frequently lag behind.
Metabolic Network Models
Cellular network models are well suited to guide process development for several reasons:
- They provide a mechanistic description of cell physiology,
- Genome-based model versions provide a direct link between cellular genotype and observed phenotypic behavior,
- They permit integration of data types such as transcript and metabolite measurements with process variables like feed rates and media composition,
- Most importantly, their mechanistic nature enables manufacturers to predict the impact of process changes on fermentation outcomes or to track down reasons why certain fermentation runs succeed while others fail.
Metabolic network models add value to metabolomics data from industrial fermentations by enabling such predictions. Model simulations support rapid hypothesis testing to assess, e.g., the impact of media changes on growth, product formation, or on intracellular metabolism.
In this article, we illustrate the application of network model-driven process design for predicting optimized media compositions for improved product titer in a CHO cell culture process.
CHO Metabolic Network Reconstruction
The metabolic network of the CHO cell was reconstructed using information from public databases and primary literature. Stoichiometry, reversibility, and the elemental composition of metabolites was modeled for every reaction. Enzymatic reactions were mapped to corresponding genes based on a genome annotation for mouse.
The model comprises different cellular compartments (cytosol, mitochondria, ER, Golgi) and the fermenter. It accounts for the glycoform structure and amino acid composition of the product molecule as well as for known cell line specific genetic modifications of metabolic enzymes.
First, we applied the model to characterize the reference process based on time series of extracellular metabolite concentrations from a fed-batch fermentation. For each physiologically distinct process phase, cell-specific rates of nutrient uptake, growth, and product formation were calculated from mass balances (Figure 1), employing Monte Carlo simulation to calculate error propagation.
From these extracellular rates, we computed intracellular flux distributions using metabolic flux analysis and the CHO network model. By comparing the flux distributions in different process phases, it is possible to identify (i) pathways with significant flux changes during the fermentation and (ii) process phases with favorable flux distribution.
Such information cannot be inferred from measured concentration time series alone, but requires the use of an appropriate model. Moreover, the results obtained provide a first assessment of cellular energy status and of nutrient fractions supplied by cellular uptake and/or synthesis, respectively.
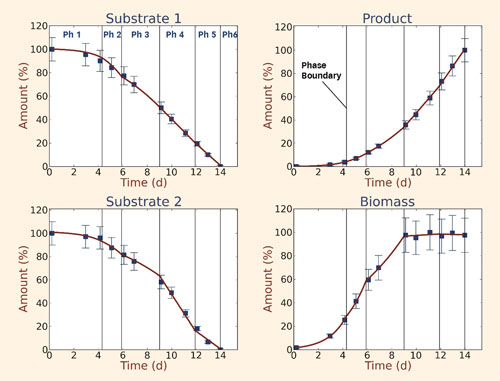
Figure 1. Time courses of extracellular metabolites and biomass during the process: Specific rates and process phases were determined from the total set of concentration time series and feed specifications using Monte Carlo simulation to calculate error propagation. Vertical lines demarcate boundaries between distinct process phases.
Optimizing Feed Media Compositions
In practice, candidate production clones for scale-up often differ in metabolic phenotype resulting in different nutrient requirements during the process. Standardized feed media compositions cannot optimally support the individual nutrient demand, which can limit achievable product titers. Comprehensive metabolic characterization of the respective process not only furthers mechanistic understanding but also represents a good starting point for rational media optimization.
We computed optimized media compositions for distinct process phases based on the observed clone specific flux distributions by combining stationary and dynamic model simulations. For dynamic simulations, uniform kinetic rate laws were assigned to individual reaction steps. Additionally, feeds and sampling were accounted for. In this way, the impact of modifications in medium composition or feed rates on product synthesis, byproduct formation, and intracellular metabolism could be predicted (Figure 2).
Model parameters were determined using evolutionary strategies on high-performance computing clusters paying attention to reaction directionality where appropriate. The resulting model served to predict feed media compositions with optimized concentrations of glucose and amino acids for two sequential feeding streams.
Application of the predicted optimized feed in the cell culture process increased final product titer by more than 50% and increased the integral of viable cells already in the first iteration (Figure 3).
Moreover, a significant reduction in ammonium formation was observed. If necessary, the procedure can be repeated to further refine media compositions. Data from replicate fermentations can be included to increase the robustness of feed media proposals but is not essential.
Due its mechanistic nature, the stoichiometric network model captures forced couplings between substrate uptake and formation of byproducts. By comparison, DoE approaches entail a much larger number of experiments to extract such information, thus requiring more time and effort.
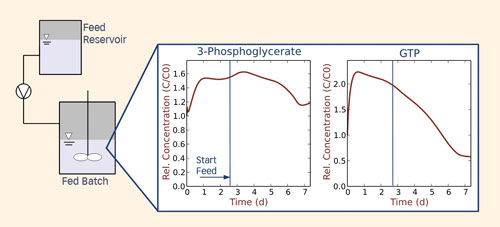
Figure 2. Simulated response of intracellular metabolite dynamics in response to feeding simulations. The applied kinetic model is based on a clone-specific flux distribution observed in vivo.
Refined Predictions & Future Prospects
The influence of different environmental conditions on the outcome of a cell culture process can already be predicted quite well using the presented approach. Our model-based approach currently focuses on predictions for amino acids and carbon sources like glucose and pyruvate.
Extension to further compounds is straightforward as long as one can reliably quantify their uptake or production. Fluxes through alternative metabolic pathways and metabolic cycles can, however, not be estimated by mass balancing alone. These parts of the metabolic network model can be quantified using 13C tracer experiments in combination with transient 13C metabolic flux analysis. This method is also applicable to industrial fed-batch processes.
Including this additional information in the metabolic network model can significantly improve the predictions. Further refinement of the model’s predictive capabilities is possible by incorporating intracellular metabolite measurements and by accurate knowledge of the cellular composition, e.g., of total protein content.
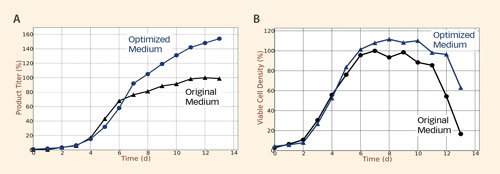
Figure 3. Product titer (A) and viable cell density (B) for CHO cells grown in the original medium or in the optimized medium that was designed by performing in silico simulations using the metabolic network model.
Conclusions
Combining metabolomics data with validated metabolic network models has great potential for adding value to data collections acquired in PAT and QbD programs. It converts metabolomic data into a key asset for (i) quantitative predictions of fermentations outcomes, (ii) accelerated rational process optimization, and (iii) improved process understanding. Possible applications range from cell-line engineering to characterization and design of production-scale processes:
- Engineering of cell lines for debottlenecking of growth and product formation through incorporation of intracellular metabolite measurements and other omics data (protein, transcript),
- Selection of robust clones with high specific productivity for scale-up based on a comprehensive metabolic characterization at small volumes,
- Derivation of media compositions tailored to clone-specific nutrient demand at reduced experimental effort compared to DoE approaches,
- Prediction of future nutrient requirements from current process status for controlling feed additions in production fermenters.
The present approach is readily transferable to other mammalian cell lines and is applicable to microbial fermentation processes as well.
Dirk Müller, Ph.D. ([email protected]), is head of biopharmaceutical services, Florian Kirchner is head of software engineering, Stefanie Mangin and Jens Niklas are project managers, and Klaus Mauch is CEO at Insilico Biotechnology. Web: www.insilico-biotechnology.com.



