Rating: Very Good
Strong Points: Features are intuitive and easy to use.
Weak Points: New tool, unclear how extensively it’s been tested by other groups.
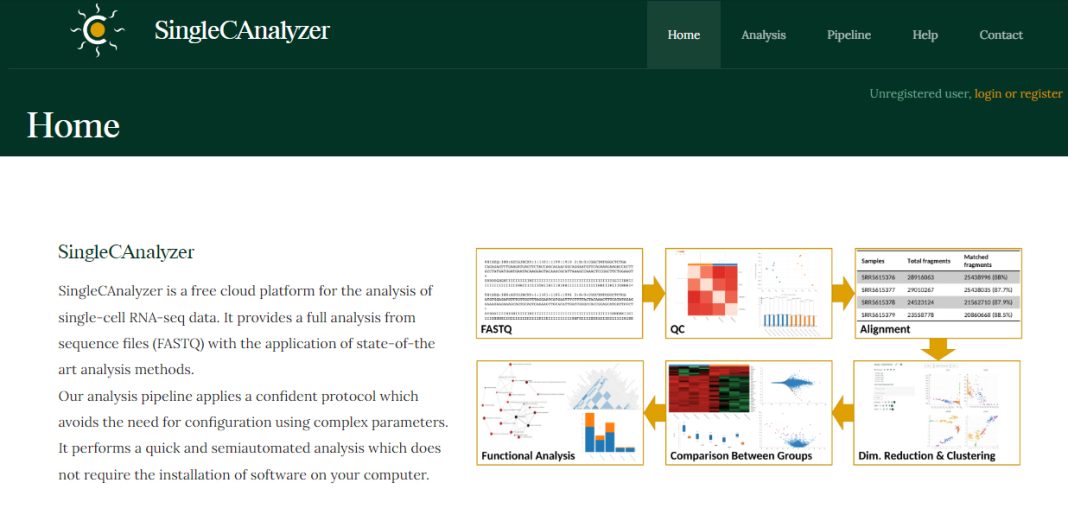
Single-cell RNA sequencing is an incredible tool that gives us unprecedented insight into transcriptomes at the smallest scale. However, with the fine resolution of this technique comes unwieldy datasets that can be difficult to parse through. Fortunately, we have tools like Single C Analyzer to make that job easier. Single C Analyzer is an open-source tool that you can run right from your browser. After uploading their sequencing file, users are walked through a pipeline that will align and trim your reads and can perform quality control, feature selection, and dimensional reduction on your data. Single C Analyzer’s built-in pipeline even allows you to perform unsupervised clustering on your data and compare samples for expression and gene enrichment. Most aspects of the pipeline are customizable—for example, when applying dimensionality reduction to your data you can choose whether to use t-SNE, UMAP, or principal components. The interface is very intuitive, but if you need some help, the website provides a tutorial video and a detailed flowchart of the full analysis pipeline.