August 1, 2015 (Vol. 35, No. 14)
Thanks to Advances in Genomics, PCR Is Becoming More Common in Clinical Applications
Since its invention by Kary B. Mullis in 1985, the polymerase chain reaction (PCR) has become well established, even routine, in research laboratories. And now PCR is becoming more common in clinical applications, thanks to advances in genomics and the evolution of more sensitive quantitative PCR methodologies.
To kick off the event, speakers shared their views on POC molecular tests. These tests, the speakers insisted, can provide significant value to healthcare only if they support timely decision making.
Clinic-ready PCR platforms need to combine speed, ease of use, and accuracy. One such platform, the cobas Liat (“laboratory in a tube”), is manufactured by Roche Molecular Systems. The system employs nucleic acid purification and state-of-art PCR-based assay chemistry to enable POC sites to rapidly provide lab-quality results.
The cobas Liat Strep A Assay detects Streptococcus pyogenes (group A β-hemolytic streptococcus) DNA by targeting a segment of the S. pyogenes genome. The operator transfers an aliquot of a throat swab sample in Amies medium into a cobas Liat Strep A Assay tube, scans the relevant tube and sample identification barcodes, and then inserts the tube into the analyzer for automated processing and result interpretation. No other operator intervention or interpretation is required. Results are ready in approximately 15 minutes.
According to Shuqi Chen, Ph.D., vp of Point-of-Care R&D at Roche Molecular Systems, clinical studies of the cobas Liat Strep A Assay demonstrated 97.7% sensitivity when the test was used at CLIA-waived, intended-use sites, such as physicians’ offices. In comparison, rapid antigen tests and diagnostic culture have sensitivities of 70% and 81%, respectively (according to a 2009 study Tanz et al. in Pediatrics).
The cobas Liat assay preserved the same ease-of-use and rapid turnaround as the rapid antigen tests. It addition, it provided significantly faster turnaround than the lab-based culture test, which can take 24–48 hours.
A CLIA waiver was announced for the cobas Liat Strep A assay in May 2015. CLIA wavers have been submitted for cobas Liat flu assays, and Roche intends to extend the assay menu.
POC tests are also moving into field applications. Coyote Bioscience has developed a novel method for one-step gene testing without nucleic acid extraction that can be as fast as 10 minutes from blood sample to result. Their portable devices for molecular diagnostics can be used as genetic biosensors to bring complex clinical testing directly to the patient.
“Instead of sequential steps, reactions happen in parallel, significantly reducing analysis time. Buffer, enzyme, and temperature profiles are optimized to maximize sensitivity,” explained Sabrina Li, CEO, Coyote Bioscience. “Both RNA and DNA can be analyzed simultaneously from a drop of blood in the same reaction.”
The first-generation Mini-8 system was used for Ebola detection in Africa where close to 600 samples were tested with 98.8% sensitivity. Recently in China, the Mini-8 system was applied in hospitals and small community clinics for hepatitis B and C and Bunia virus detection. The second-generation InstantGene system is currently being tested internally with clinical samples.
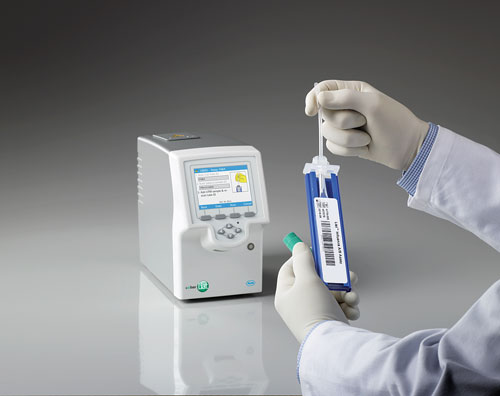
Last May, Roche Molecular Systems announced that its cobas Liat Strep A assay received a CLIA waiver. This clinic-ready assay can detect Streptococcus pyogenes (group A ß-hemolytic streptococcus) DNA in throat swabs by targeting a segment of the S. pyogenes genome.
Digital PCR
Personalized cancer care requires ultra-sensitive detection and monitoring of actionable mutations from patient samples. The high sensitivity and precision of droplet-digital PCR (ddPCR) from Bio-Rad Laboratories offers critical advantages when clinical samples are limited, degraded, or contain PCR inhibitors.
Typically, formalin-fixed and paraffin-embedded (FFPE) tissue samples are processed. FFPE samples work well for immunohistochemistry and protein analysis; however, the formalin fixation can damage nucleic acids and inhibit the PCR reaction. Samples may yield 100 ng of purified nucleic acid, but the actual amplifiable material is less than 1%, or 1 ng, in most cases.
“Current qPCR technology depends on real-time fluorescence accumulation as the PCR is occurring, which can be an effective means of detecting and quantifying DNA targets in nondegraded samples,” commented Dawne Shelton, Ph.D., staff scientist, Digital Biology Center, Applications Development Group, Bio-Rad Laboratories. “Amplification efficiency is critical; if that amplification efficiency changes because of sample quality it is hidden in the qPCR methodology.”
“In ddPCR, that is a big red flag,” Dr. Shelton continued. “It changes the format of how the data look immediately so you know the amount of inhibition and which samples are too inhibited to use.”
Tissue types vary and contain different degrees of fat or other content that can also act as PCR inhibitors. In blood monitoring, the small circulating fragments of DNA are extremely degraded; in addition, food, supplements, or other compounds ingested by the patient may have an inhibitory effect.
Clinical labs test for these variabilities and clean the blood, but remnant PCR inhibitors can remain. In ddPCR, a single template is partitioned into a droplet. If the droplet contains a good template, it produces a signal; otherwise, it does not—a simple yes or no answer.
“Even if there is no PCR inhibition, most clinical samples yield very small amounts of nucleic acid,” Dr. Shelton added. “To make a secure decision using qPCR is difficult because you are in a gray zone at the very end of its linear range. ddPCR operates best with small sample amounts and provides good statistics for confidence in your results.”
Currently, at least a half dozen clinical trials worldwide are using digital PCR, half of them are using the Bio-Rad QX200 Droplet Digital PCR system. Examples of studies include examining BCR-ABL monitoring in patients with chronic myelogenous leukemia (CML); identifying activating mutations in epidermal growth factor receptor (EGFR) for first-line therapy of new drugs in patients with lung cancer; and the monitoring of resistance mutations such as EFGR T790M in patients with non-small cell lung cancer (NSCLC).
Clovis Oncology used a technology called BEAMing (Beads, Emulsions, Amplification, and Magnetics), a type of digital PCR for blood-based molecular testing, to perform EGFR testing on almost 250 patients in clinical trials. In BEAMing, individual EGFR gene copies from plasma are separated into individual water droplets in a water-in-oil emulsion. The gene copies are then amplified by PCR on magnetic beads.
The beads are counted by flow cytometry using fluorescently labeled probes to distinguish mutant beads from wild-type. Because each bead can be traced to an individual EGFR molecule in the patient’s plasma, the method is highly quantitative.
“BEAMing is particularly well-suited for the detection of known mutations in circulating tumor DNA. In this circumstance, the mutation of interest often occurs at low levels, perhaps only 1–2 copies per milliliter or even less, and in a high background of wild-type DNA that comes from normal tissue. BEAMing can detect one mutant molecule in a background of 5,000 wild-type molecules in clinical samples,” stated Andrew Allen, MRCP, Ph.D., chief medical officer, Clovis Oncology.
In the studies, the EGFR-resistance mutation T790M could be identified in plasma 81% of the time that it was seen in the matched patient tumor biopsy. Additionally, about 10% of patients in the study had a T790M mutation in plasma that was not identified in tissue, presumably because of tumor heterogeneity. Another 5–10% of the patients did not provide an EGFR result, usually because the tissue biopsy had no tumor cells.
In aggregate, these results suggest that plasma EGFR testing can be a valuable complement to tumor testing in the clinical management of NSCLC patients, and can provide an alternative when a biopsy is not available. Tumor biopsies may provide only limited tissue, if in fact any tissue is available, for molecular analysis. Also, mutations may be missed due to tumor heterogeneity. These mutations may be captured by sampling the blood, which acts as a reservoir for mutations from all parts of a patient’s tumor burden.
In the last few years, a panoply of clinically actionable driver mutations have been identified for NSCLC, including mutations in EGFR, BRAF, and HER2, as well as ALK, ROS, and RET rearrangements. These driver mutations will migrate NSCLC molecular diagnostic testing in the next few years toward panel testing of relevant cancer genes using various digital technologies, including next-generation sequencing.
Conventional real-time PCR technology, while suited to the analysis of high-quality clinical samples, may effectively conceal amplification efficiency changes when sample quality is inconsistent. A more effective alternative, Bio-Rad suggests, is its droplet-digital PCR (ddPCR) technology, which can provide absolute quantification of target DNA or RNA, a critical advantage when samples are limited, degraded, or contain PCR inhibitors. The company says that of the half-dozen clinical trials that are using digital PCR, half rely on the Bio-Rad QX200 ddPCR system.
SNP Analysis
The Rapid Cycle In-line PCR instrument, a research-use-only platform from Canon U.S. Life Sciences, has been shown to streamline testing of multiple single nucleotide polymorphisms (SNPs) on a set of genomic DNA samples. By utilizing liquid-handling robots, the instrument automates serial PCR reaction preparation and dispensation to a microfluidic-based reaction cartridge, decreasing hands-on time and potential handling errors. The small reaction volumes significantly reduce the time required to complete PCR amplification and DNA-melting analysis.
“To date, the instrument has been used successfully in genotyping SNPs, and we are investigating other genetic targets including, but not limited to, small insertions and deletions,” said Hanyoup Kim, Ph.D., senior scientist, Canon U.S. Life Sciences.
In contrast to PCR assays for gene expression, SNP genotyping assays must precisely target a specific position within a gene. This spatial restriction limits the nucleotide sequences that PCR primers and/or probes will target, and in turn this impacts the cycling parameters (annealing temperatures and times) that will yield robust amplification. Therefore, users will often require different PCR conditions to effectively test all of their loci of interest.
Lot-to-Lot Consistency
PCR-based molecular diagnostic assays rely on high-quality reaction components to yield robust, reliable, and reproducible results. Assay components must perform the same, lot to lot.
For clinical applications that require additional assurances, Promega has added multiple levels of control to their processes. The manufacturing process and quality control methods are validated and supplied with appropriate documentation to demonstrate repeated supply of a consistent product. Robust assays test for factors that can negatively impact an assay, such as DNA contamination, enzyme concentration, and activity. Detailed certificates of analyses dovetail with users’ quality systems.
“We have custom manufacturing abilities that can adapt to the specific needs of clinical customers and can involve anything from format to very specific formulations or bulk product manufacture,” asserted Terri McDonnell, senior program manager, Promega. “Customers may need a specific enzyme concentration, formulation, or volume; custom documentation; [or] a specific QC assay—or [they may] even [need] help on assay design.”
“Building upon the base of controls that we have in place around the manufacture of our products is really important for our molecular-diagnostic customers,” McDonnell continued. “It gives us the ability to customize products while maintaining control.”
As assays are being pushed out closer to POC, such as in field work, temperature and humidity control can be a problem. Reagents need to be stable under very diverse conditions, and requests are growing more frequent for glycerol-free reagents that can be lyophilized. In the future, as the use of more complex multiplexed assays and panel testing becomes more common, lot-to-lot consistency along with supporting documentation will become more important.

Promega, which produces reaction components for PCR-based molecular assays, notes that the quality controls implemented by manufacturers can affect molecular diagnostic assay design and performance. The company adds that as multiplexed assays and panel tests become more common, lot-to-lot reagent consistency will become more important.
Live Sentinel Mice vs. PCR Testing
Most lab animal facilities monitor mouse colonies for pathogens that can negatively impact research. The current health monitoring program at Oregon Health and Science University (OHSU) consists of exposing three sentinel mice to soiled bedding from 70 colony cages during biweekly cage changes.
Hoping to improve detection methods and reduce the number of mice necessary for sentinel testing, OHSU, working with Charles River Laboratories, decided to evaluate whether PCR testing using swabs collected from a single location on the exhaust plenum of each individually ventilated cage rack would be equivalent to results obtained from testing sentinel animals.
The team’s results demonstrated that Exhaust Air Dust (EAD) sampling is a viable option for collecting samples and obtaining results indirectly from colony animals, according to Kim Saunders, D.V.M., who heads OHSU’s department of comparative medicine.
“Eliminating the use of live sentinel mice would result in a substantial savings in costs for monitoring the health of colony animals,” says Dr. Saunders. “EAD testing appears to have great promise in our goal of refinement, reduction, and replacement of live animals and could potentially save thousands of rodents’ lives.”
Ken Henderson, Ph.D., director of R&D at Charles River Labs, also agrees that EAD sampling appears to be cost-effective. “Yearly costs for the Oregon sentinel program ran around $191,000 compared to around $27,000 for EAD samples without the use of live sentinels,” he says.



