April 1, 2013 (Vol. 33, No. 7)
As the sequence of DNA molecules is so fundamental to the understanding and treatment of diseases, the efforts to decode this sequence in humans has been identified as a grand challenge over the last three decades.
Since the completion of the Human Genome Project in the mid 2000s, the cost of decoding the human genome has dramatically fallen from $100 million per genome to now less than $5,000. The cost could perhaps be driven down even further due to the development of single molecule nanopore sensors that promise the direct, label- and amplification-free detection of DNA sequences.
The operating principle of a nanopore sensor is similar to that of a Coulter counter. A single-charged DNA molecule is electrophoretically driven through a nanopore under an applied electric potential, where the pore is formed in an insulating membrane separating two chambers filled with electrolyte solution. An electrical current is measured due to the movement of the ions through the nanopore.
When the DNA enters and passes through the pore, the ionic current generally decreases and the current pulse-width and depth can reveal useful information about the length and structure of the DNA. The application of nanopores to DNA sequencing was first described by Church, Deamer, Branton, Baldarelli, and Kasianowicz in a patent application that was filed in 1995 and awarded in 1998. The first experimental verification of single DNA movement in nanopore was reported by Kasianowicz, et al., at NIST in 1996.
Since then, the holy grail has been to measure the ionic current with high enough spatial and temporal sensitivity to see different electrical signature when each nucleotide or base passes through the nanopore. Hence, by measuring the ionic current through the nanopore over time, it could be possible to directly “read” the sequence of bases in the DNA.
This is indeed a very attractive proposition. However two main challenges remain: slowing down the molecule through the nanopore and having a high enough spatial resolution to allow single-base detection.
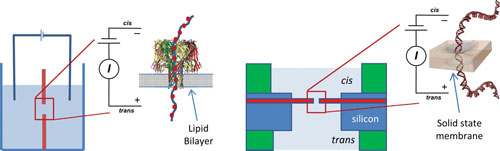
Schematic representation of a biological nanopore in a lipid bilayer (left) and a nanopore in a solid state membrane (right) statistically significant admixture.
Categories Under Investigation
To date, two main nanopore categories are under investigation: biological nanopores and solid-state nanopores. Biological nanopores, which are inserted into a lipid bilayer, can be produced in large numbers with a size precision unmatched by the semiconductor industry today.
The most commonly reported biological nanopore is the heptameric protein α-hemolysin, which has a transmembrane β-barrel that is ~2.6 nm wide and ~5 nm long, with the narrowest point being just 1.4 nm wide. This allows single-stranded DNA to translocate (but not double-stranded DNA). This channel was used in the first experimental demonstration and still remains a choice for next-gen DNA sequencing.
One particular challenge is the electrophoretic speed of DNA, which is on the order of 1 base per microsecond, making it nearly impossible to perform low noise, low current electrical measurements to sequence freely translocating ssDNA with α-hemolysin.
A promising approach to regulating the speed of DNA through the nanopores is via the use of enzymes such as DNA polymerases, which are coupled with the biological nanopores. Taking this approach, the individual catalytic cycles can be detected with the corresponding perturbations in the ionic current measurements in the nanopores.
Another related approach developed by Haygan Bayley of Oxford University is that of “sequencing by digestion.” In this technique, an exonuclease is chemically coupled to an α-hemolysin anchored in a lipid bilayer. The exonuclease cleaves off individual bases from a DNA molecule, and the bases are then identified by differences in the ionic current as they pass through a mutant α-hemolysin channel with a molecular recognition element inside the channel.
The technology is licensed to Oxford Nanopore Technology for commercialization as a next-generation sequencing approach.
The octameric protein mspA, which has a channel that is just ~0.5 nm long with a constriction of diameter of ~1.2 nm at the narrowest point, represents yet one more attractive biological nanopore channel. mspA can detect as few as three nucleotides contributing to the pore current compared with the ten or so nucleotides that change the current in the α-hemolysin described previously.
Again, without any processive enzymes, the DNA molecules move too fast to perform a direct reading of the sequence. Hence enzymes such as the phi29 DNA polymerase could be coupled to slow the molecule.
Techniques such as duplex-interrupted nanopore sequencing might be able to overcome the issue of high translocation speeds by inserting a “short” segment of dsDNA between each nucleotide in an analyte DNA molecule. This can momentarily halt the DNA molecule allowing single nucleotides to be identified by perturbations in the ionic current.
Significant Advancements
Although significant advancements have been made with biological nanopores toward next-generation sequencing, the first commercial nanopore approaches will most likely utilize biological channels. But solid-state counterparts also pose attractive possibilities.
The solid-state membrane made of silicon nitride or other material can replace the lipid bilayer, and a focused electron beam is used to drill a nanometer scale pore through the membrane. Since the first report in 2001 by Golovchenko, many groups have reported forming pores and translocation of DNA under applied electric fields.
The solid-state pores possess superior mechanical and thermal characteristics compared to their biological counterparts. These devices have been used to study the fundamental biophysical phenomenon of molecules moving through nanopores, the detection of nucleic acid molecules such as miRNA, epigenetics for diagnostic applications and, of course, the quest for direct label-free detection of DNA sequences.
This approach, however, requires the ability to form ultrathin membranes and narrow pores comparable to the size of single nucleotides. Atomic layer deposition of solid-state materials is an attractive option to make thin membranes, as it can allow for precise control of membrane thickness and material composition. It has been used to make nanopores in aluminum oxide.
However, the goal to make a membrane thin enough to allow the interrogation of a single or just a few bases remained a challenge until the last few years.
Recent developments in the exploration of graphene materials offer exciting possibilities. Along with its unique physical structure and high electronic mobility, this two-dimensional sheet of carbon atoms has a thickness that is comparable to the spacing between nucleotides in ssDNA (0.32–0.52 nm). As a result, the use of graphene as a membrane is particularly attractive for electronic DNA sequencing.
Drndic and co-workers first demonstrated the fabrication of nanopores in suspended graphene films. Subsequently, groups led by Golovchenko, Drndic, and Dekker reported the DNA translocation through nanopores made of graphene. This field is quite nascent with only a handful of studies having been reported to date.
As graphene is electrically conducting in the plane of the 2D lattice, these ultrathin electrodes could be used to provide new device and sensing topologies such as graphene nano ribbons and graphene-based tunnel junctions. The speed of the molecule is still too fast and methods to slow down the molecule are being pursued.
The use of nanopore-based sensors to perform long base reads on unlabeled, single-stranded DNA molecules quickly and cheaply can revolutionize the fields of genomics and personalized medicine. Many challenges in sequencing with biological nanopores, such as the high translocation velocity and the lack of nucleotide specificity, already have been addressed and commercially viable approaches are being pursued.
Solid-state counterparts also promise to evolve into robust methodologies although the challenges of speed and specificity still exist. If the speed of the molecules could be reduced to a single nucleotide per millisecond, thus allowing high fidelity electronic measurements of single bases, and if nucleotides could have a unique electronic signature, then it might be possible to sequence molecules containing one million bases in less than 20 minutes.
The outlook for developing such nanopore-based next-gen sequencing tools remains bright and there is little doubt that nanopore-based sensors will continue to develop as strong candidates to join other third-generation sequencing technologies in the race toward low-cost personalized DNA sequencing.
$3M Prize to Fund Telomeres Research
Rosalind Franklin Society board member Titia de Lange, Ph.D., director of the Anderson Center for Cancer Research, Leon Hess Professor and head of the Laboratory of Cell Biology and Genetics at the Rockefeller University received a Breakthrough Prize in Life Sciences.
The Breakthrough Prize in Life Sciences, founded by the chairman of Apple and Genentech Art Levinson, Google co-founder Sergey Brin, 23andMe founder Anne Wojcicki, Facebook founder Mark Zuckerberg and his wife Priscilla Chan, and Russian entrepreneur and philanthropist Yuri Milner, awarded 11 scientists with $3 million each to recognize their excellence in research aimed at curing intractable diseases and extending human life.
Dr. de Lange’s award will fund her research on telomeres. The de Lange lab identified a six-subunit protein complex, which they named shelterin, that specifically binds to telomeres. Dr. de Lange and her colleagues determined the fate of telomeres lacking one or more of the six shelterin subunits by using Cre-mediated conditional deletion in mouse embryo fibroblasts. According to their research, cells lacking shelterin perceive their natural chromosome ends as sites of DNA damage. Her group is now working to determine the mechanism by which each shelterin protein inhibits its designated pathway, and how loss of telomere protection contributes to genome instability in human cancer.
Dr. de Lange, along with the other winners, will be invited to present their findings in public lectures and have agreed to serve on the selection committee of the foundation to choose recipients of future prizes. Dr. de Lange and Cori Bargmann, Ph.D., Investigator, Howard Hughes Medical Institute and Torsten N. Wiesel professor at the Rockefeller University, are the only women chosen to receive this prize.
The Rosalind Franklin Society recognizes and advances the significant contributions of women in the life sciences and affiliated disciplines. The society works to enable women to achieve more tenure-track appointments in academia as well as leadership positions in industry and government.
Rashid Bashir, Ph.D. ([email protected]), is the Abel Bliss professor of engineering in the department of bioengineering and electrical and computer engineering at the University of Illinois-Urbana-Champaign.



