Colorectal cancer (CRC) remains the fourth leading cause of cancer death worldwide, and while the introduction of widespread screening has reduced mortality, current screening techniques have their drawbacks.
While considered the gold standard for detecting precancerous polyps, colonoscopies are invasive procedures, and many people avoid or delay getting screened for CRC this way. About seven in 10 U.S. adults between 50 and 75 are up to date with recommended colonoscopy screenings.
Take-home tests such as FIT and Cologuard that look for signs of shed tumor DNA in stool samples are noninvasive but less accurate.
“Cell-free DNA is quickly broken down and degraded in the intestinal lumen,” said Simon Leedham, PhD, a professor of molecular and population genetics at the University of Oxford.
But a new study from researchers at the University of California, San Diego (UCSD), and multiple Australian institutions published in Science hints at the possibility of new tools for detecting colorectal and intestinal cancers: bioengineered bacteria that can detect cell-free DNA shed by tumors in close proximity to those tumors, while still inside the digestive tract.
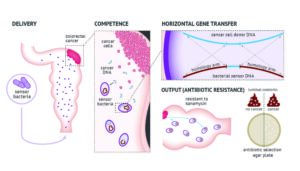
“Bacteria that have integrated the freshly shed DNA should allow an increase in sensitivity of detection that will hopefully outperform existing tools that look for the presence of microscopic quantities of blood or shed (epi)genetic mutations in the stool,” said Leedham, who is familiar with but not involved in the new research. “This will be key for also detecting the precursors to cancers, intestinal polyps, which can be safely removed endoscopically and prevent patients from going on to develop cancer in the first place.”
The researchers chose the Acinetobacter baylyi bacterium, known for its facility at natural competence, that is, the ability to take up genetic material from the environment and integrate it into its own genome through homologous recombination, according to Robert Cooper, PhD, a scientist at the UCSD Synthetic Biology Institute and lead author of the new paper.
“A. baylyi is not the only species that does this, but they are really good at it,” he added. “If there was a closely related gene floating around that’s similar to one of their genes they could use that homologous recombination to replace theirs with a similar version.“
A role for CRISPR-Cas9
To take advantage of this natural propensity, Cooper and his colleagues used CRISPR-Cas9 to insert a mutated version of the human KRAS oncogene, a form of the gene found in 27% of colorectal cancers. They also inserted the KanR gene, which provides bacteria with resistance to the antibacterial agent kanamycin. When the engineered bacteria integrate the tumor KRAS gene, KanR is activated, with the acquired kanamycin resistance serving as the output signal.
The way this worked in the study is that the researchers first inserted engineered colorectal cancer organoids into mice. They then introduced the engineered bacteria rectally and then sacrificed the mice to acquire cultures from their colons. They then spread the culture on an agar plate treated with antibiotics—the bacteria that positively identified and integrated the mutated KRAS genes shed by the organoids flourished thanks to their acquired resistance.
“That’s not what you would use in a clinical setting,” Cooper noted, “but there are systems other people have worked out to create outputs. You could possibly get urine or blood samples, or stool samples.”
There are some limitations to the technique. First, it may be limited to cancers of the digestive tract, according to Cooper.
”[The digestive track is] bathed in bacteria already and the tumors would be growing right at the interface between the body and where the bacteria are, releasing DNA into that environment where they can be sampled,” he said.
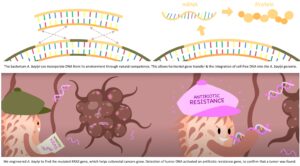
Additionally, the strategy only works for mutations that have already been defined. Not all tumors have the same mutations, and some cancers manifest a wide array of mutations, but “it happens that colorectal cancer is one of those that has a small set of very common mutations,” said Cooper.
The technique must also translate to humans, though Cooper pointed out, “This is one of those cases where scaling up to humans might actually help it because everything is bigger, so there’s more target DNA and more biosensors you can fit into the gut.”
If further developed and approved for human use, the engineered bacterial biosensors could be used for other applications, such as rapid diagnosis of gut infections. Additionally, shifting from a detection model to a therapeutic mode, the same bacteria could be engineered to release anti-tumor agents upon detecting tumor DNA, rather than sending a signal that they’ve detected it.
”That putout gene could not just be a signal, but a therapeutic molecule,” Cooper said. “With living biosensors, you can make them make whatever you want so long as it is a biological [that can be made] with a relatively easy pathway.”
That long-term possibility is what most excites Leedham about the new work. “Perhaps the near future portends the development of cancer cell ‘seek and destroy’ bacterial therapeutics,” he said.


