![Restriction enzymes are essential tools for recombinant DNA technology that have revolutionized modern biological research; however, they have limited sequence specificity and availability. The <i>Pyrococcus furiosus</i> Argonaute (PfAgo)-based platform for generating artificial restriction enzymes (AREs) is capable of recognizing and cleaving DNA sequences at virtually any arbitrary site and generating defined sticky ends of varying length. [Behnam Enghiad and Huimin Zhao/University of Illinois at Urbana-Champaign]” /><br />
<span class=](https://genengnews.com/wp-content/uploads/2018/08/Feb10_2017_UnivIllinois_RestrictionEnzymes5311545651-1.jpg) Restriction enzymes are essential tools for recombinant DNA technology that have revolutionized modern biological research; however, they have limited sequence specificity and availability. The Pyrococcus furiosus Argonaute (PfAgo)-based platform for generating artificial restriction enzymes (AREs) is capable of recognizing and cleaving DNA sequences at virtually any arbitrary site and generating defined sticky ends of varying length. [Behnam Enghiad and Huimin Zhao/University of Illinois at Urbana-Champaign]
Restriction enzymes are essential tools for recombinant DNA technology that have revolutionized modern biological research; however, they have limited sequence specificity and availability. The Pyrococcus furiosus Argonaute (PfAgo)-based platform for generating artificial restriction enzymes (AREs) is capable of recognizing and cleaving DNA sequences at virtually any arbitrary site and generating defined sticky ends of varying length. [Behnam Enghiad and Huimin Zhao/University of Illinois at Urbana-Champaign]
Scientists at the University of Illinois say they have developed a new technique of genetic engineering for basic and applied biological research and medicine. Their work (“Programmable DNA-Guided Artificial Restriction Enzymes”), reported in ACS Synthetic Biology, could open new doors in genomic research by improving the precision and adherence of sliced DNA, according to the investigators.
“Using our technology, we can create highly active artificial restriction enzymes with virtually any sequence specificity and defined sticky ends of varying length,” said Huimin Zhao, Ph.,D., professor of chemical and biomolecular engineering, who leads a synthetic biology research group at the Carl R. Woese Institute for Genomic Biology at Illinois. “This is a rare example in biotechnology where a desired biological function or reagent can be readily and precisely designed in a rational manner.”
Restriction enzymes cut DNA at a specific site and create a space wherein foreign DNA can be introduced for gene-editing purposes. This process is not achieved only by naturally occurring restriction enzymes; artificial restriction enzymes, or AREs, have risen to prominence in recent years. CRISPR/Cas9, a bacterial immune system used for “cut-and-paste” gene editing, and TALENs, or transcription activator-like effector nucleases, which are modified restriction enzymes, are two popular examples of such techniques.
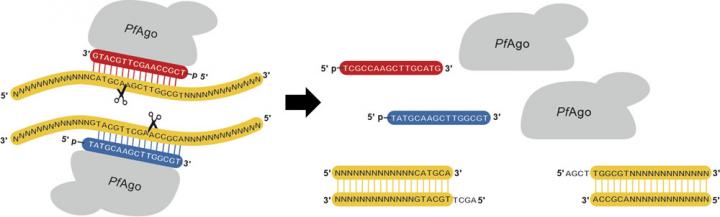
Though useful in genetic engineering, no AREs generate defined “sticky ends”—an uneven break in the DNA ladder structure that leaves complementary overhangs, improving adhesion when introducing new DNA. “If you can cleave two different DNA samples with the same restriction enzyme, the sticky ends that are generated are complementary,” explained graduate student Behnam Enghiad. “They will hybridize with each other, and if you use a ligase, you can stick them together.”
However, restriction enzymes themselves have a critical drawback: the recognition sequence that prompts them to cut is very short, usually only four to eight base pairs. Because the enzymes will cut anywhere that sequence appears, researchers rely on finding a restriction enzyme whose cut site appears only once in the genome of their organism or plasmid, an often difficult proposition when the DNA at hand might be thousands of base pairs long.
This problem has been partially solved simply by the sheer number of restriction enzymes discovered: more than 3600 have been characterized, and over 250 are commercially available. “Just in our freezer, for our other research, we have probably over 100 different restriction enzymes,” said Enghiad. “We look through them all whenever we want to assemble something. The chance of finding the unique restriction site is so low.”
“Our new technology unifies all of those restriction enzymes into a single system consisting of one protein and two DNA guides. Not only have you replaced them, but you can now target sites that no available restriction enzymes can.”
The new method creates AREs through the use of an Argonaute protein (PfAgo) taken from Pyrococcus furiosus, an archeal species. Led by a DNA guide, PfAgo is able to recognize much longer sequences when finding its cut site, increasing specificity and removing much of the obstacles posed by restriction enzymes. Furthermore, PfAgo can create longer sticky ends than even restriction enzymes, a substantial benefit as compared to other AREs.
“When we started, I was inspired by a paper about a related protein—TtAgo. It could use a DNA guide to cleave DNA, but only up to 70 degrees,” continued Enghiad. “DNA strands start to separate over 75 degrees, which could allow a protein to create sticky ends. If there were a protein that was active at higher temperatures, I reasoned, that protein could be used as an artificial restriction enzyme. So I started looking for that, and what I found was PfAgo.”
In addition to replacing restriction enzymes in genetic engineering processes, Enghiad and Dr. Zhao believe their technology will have broad applications in the biological research. By creating arbitrary sticky ends, PfAgo could make assembly of large DNA molecules easier and would enable cloning of large DNA molecules, such as biochemical pathways and large genes.
The application of these techniques is broad-reaching, they say, ranging from discovery of new small-molecule drugs to engineering of microbial cell factories for synthesis of fuels and chemicals to molecular diagnostics of genetic diseases and pathogens, which are the areas Dr. Zhao and Enghiad are currently exploring.
“Due to its unprecedented simplicity and programmability (a single protein plus DNA guides for targeting), as well as accessibility…we expect PfAgo-based AREs will become a powerful and indispensable tool in all restriction enzyme or nuclease-enabled biotechnological applications and fundamental biological research,” predicts Dr. Zhao. “It is to molecular biology as the CRISPR technology is to cell biology.”