An unlikely collaboration between a dental and an evolutionary biologist at the University at Buffalo has uncovered a new mechanism that explains how genes acquire new functions.
Using a combination of bioinformatics, phylogenetics, proteomics, and histology, a team led by evolutionary anthropologist Omer Gokcumen, PhD, and oral biologist Stefan Ruhl, DDS, PhD, compared genomic differences in a class of sugary proteins found in mucus called mucins. They discovered 15 instances in 49 mammalian species where mucins evolved by incorporating repeats in the protein-coding regions of their genes (exons) that induce the protein product to acquire a dense brush of projecting sugar molecules upon synthesis, through a process called O-glycosylation.
Gokcumen, an associate professor of biological sciences in the University at Buffalo, College of Arts and Sciences, said, “I don’t think it was previously known that protein function can evolve this way, from a protein gaining repeated sequences. A protein that isn’t a mucin becomes a mucin just by gaining repeats. This is an important way that evolution makes slime. It’s an evolutionary trick, and we now document this happening over and over again.”
Details of the novel additive mechanism of mucin’s convergent evolution were published in the journal Science Advances (“A mechanism of gene evolution generating mucin function”). Based on their findings, the authors propose exonic repeats and variations in their numbers cause genes to acquire new functions. In the case of mucins, the authors said, the repeats are key to the proteins’ slimy property. The results suggest proteins secreted by epithelial cells that have an abundance of proline residues are natural precursors of mucins. The authors believe their findings may be particularly helpful in understanding the functional evolution of sugar-linked (glycosylated) proteins.

Ruhl, a professor of oral biology and interim dean at the University at Buffalo School of Dental Medicine, has been studying mucins in saliva for three decades, primarily focusing on their ability to protect tooth decay by balancing oral microbiota.
“The repeats we see in mucins are called ‘PTS repeats’ for their high content of the amino acids proline, threonine, and serine. They aid mucins in their important biological functions that range from lubricating and protecting tissue surfaces to helping make our food slippery so that we can swallow it,” said Ruhl. “Beneficial microbes have evolved to live on mucus-coated surfaces, while mucus can at the same time also act as a protective barrier and defend against disease by shielding us from unwanted pathogenic intruders.”
While studying saliva, the researchers noticed that MUC7, a small salivary mucin in humans, was missing in mice, a species that expresses MUC10, another mucin of comparable size. While investigating their origins, the scientists found that although the human MUC7 was not evolutionarily related to the mouse MUC10, a non-mucin protein in human tears, PROL1, shared structural domains with MUC10 but did not include the mucin’s characteristic dense brush of sugar-coated repeats.
“We think that somehow that tear gene ends up repurposed,” Gokcumen said. “It gains the repeats that give it the mucin function, and it’s now abundantly expressed in mouse and rat saliva.”
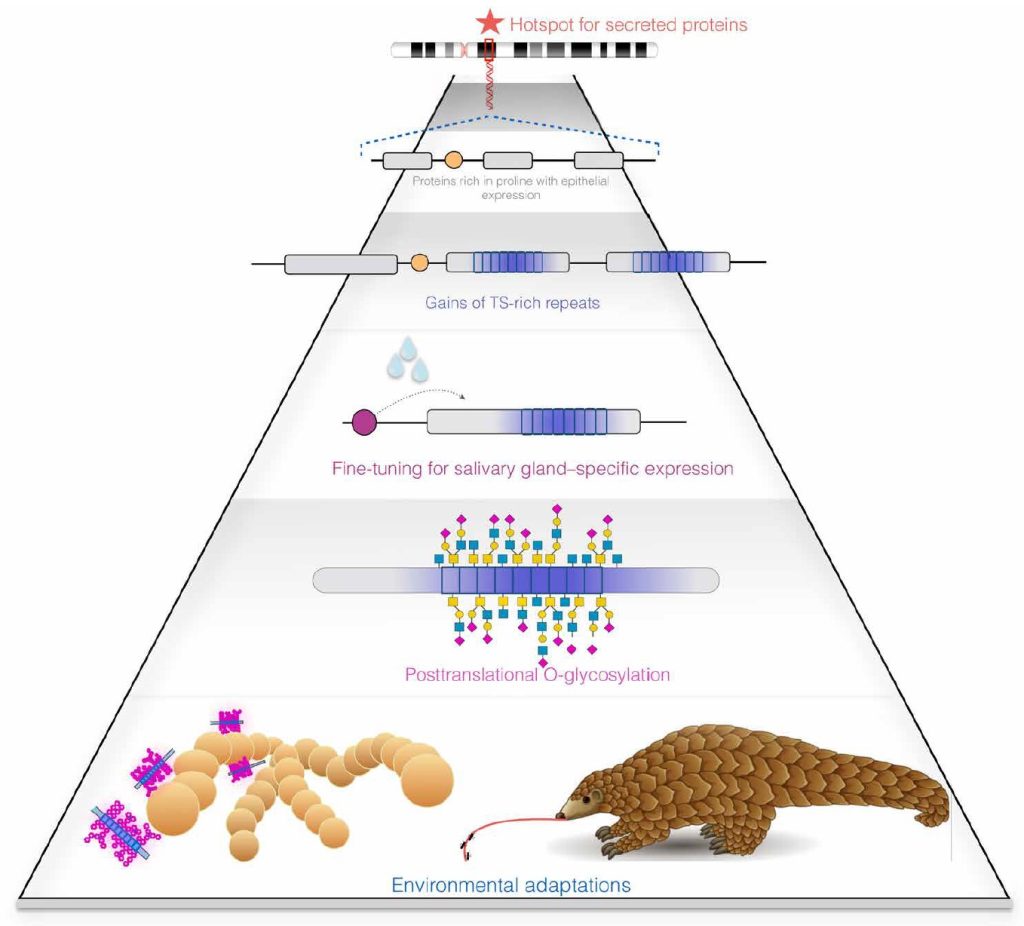
This led the team to investigate the origin of other mucins to probe whether MUC10’s “mucinization” represented a common evolutionary paradigm. They discovered 15 instances of the same phenomena that support their hypothesis that non-mucins convert to mucins by the inclusion of PTS repeats. Considering that the study focused on only one region of the genome in a few dozen mammalian species, Gokcumen said, their conclusions are “pretty conservative.” He is curious to investigate whether mucins in slugs, slime eels, and other species have arisen through the same mechanism.
First author of the study, Petar Pajic, a graduate student in Gokcumen’s lab said their findings add to the body of work that attempts to explain how gene functions evolve and may have wider ramifications.
“This could have even broader implications, both in understanding adaptive evolution and in possibly explaining certain disease-causing variants,” Pajic added. “If these mucins keep evolving from non-mucins in different species at different times, it suggests some adaptive pressure that makes it beneficial. At the other end of the spectrum, if this mechanism goes ‘off the rails’—happening too much, or in the wrong tissue—it could lead to diseases like certain cancers or mucosal illnesses.”