Ion channel–specific antigen-binding fragments (Fabs) were developed by in vitro phage display selection using Salipro®, Salipro Biotech’s proprietary nanomembrane platform technology. Isolated Fabs bound with high affinity to conformational epitopes present on the native ion channel in cell membranes.
The challenge of stabilizing complex membrane proteins
Monoclonal antibodies (mAbs) have become a prominent class of drugs during the last decade. In fact, the global therapeutic antibody market is expected to be worth $179.56 billion by 2025.1 However, a technical hurdle in the mAb discovery field is the lack of methods to routinely produce purified target antigens in clinically relevant conformations to support the discovery of target-specific binders with desired functional properties.
This is especially the case for the generation of therapeutic antibodies directed at complex, multiple transmembrane targets including G protein–coupled receptors (GPCRs), ion channels, and transporters.2,3 The untapped potential in developing mAbs toward complex multiple transmembrane targets is evident, as more than half of all clinically approved small-molecule drugs target complex membrane proteins. In comparison, only two GPCR-targeting mAbs have been approved by the U.S. Food and Drug Administration (FDA), and no mAbs targeting ion channels or transporters have been approved.4
One of the main hurdles to the development of novel therapeutic antibodies against GPCRs and ion channels is directly coupled to the transmembrane topology of these targets, making them difficult to extract and purify from the lipid bilayer of the cell without disrupting their native conformation. Detergent micelles, which are often used for membrane solubilization, cause disruption of the lipid bilayer and form artificial detergent molecular micelles that surround the transmembrane regions of the solubilized membrane protein. In the process, the membrane protein may become unstable due to the loss of its natural lipid environment.3,5,6
Several technical advances to formulating purified antigens, such as virus-like particles (VLPs), liposomes, nanodiscs, and detergent optimization, have occasionally been shown to generate promising therapeutic mAb candidates.2,5,7,8 However, it is laborious and time consuming to screen each of these platforms and adjust protocols for every antigen, and to date there is no technology platform that has been able to routinely generate therapeutic leads directed at membrane protein targets.
Salipro platform technology for stabilization of challenging drug targets
Swedish biotech company Salipro Biotech is focused on unlocking challenging drug targets for the development of next-generation therapeutics. To date, Salipro Biotech has signed multiple research collaborations with top-tier pharma and biotech companies to accelerate the discovery of novel drugs. Headquartered in Stockholm with a fully owned IP portfolio, the company has developed its platform technology for the stabilization of membrane proteins.
The Salipro technology reconstitutes challenging membrane proteins while preserving the native structure and function of these drug targets in stable, lipid-containing nanoparticles.9,10 This is achieved with the Salipro scaffold protein, which self-assembles
into disc-like nanoparticles comprised of lipids and membrane proteins (Figure 1).
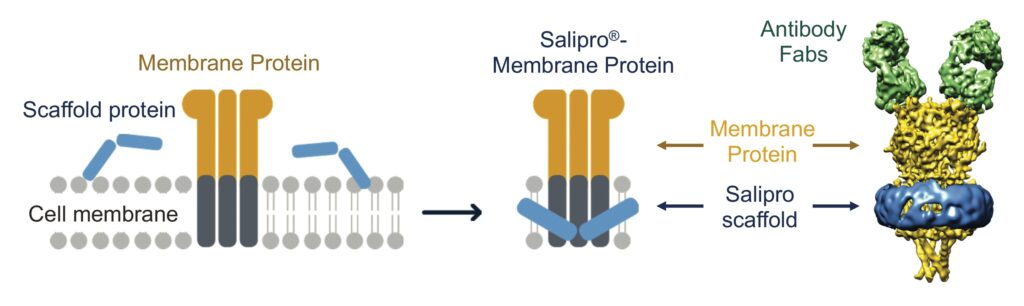
The advantage of the Salipro platform is that the scaffold flexibly adapts to the transmembrane regions of the drug target, making it possible to reconstitute membrane proteins into nanoparticles independent of size or shape. Furthermore, the Salipro scaffold allows for reconstitution of fragile multiple transmembrane targets (such as GPCRs, ion channels, and transporters) directly from crude cell membranes solely using the native lipids associated with the target (Figure 1).
Drug targets stabilized in Salipro nanoparticles can be utilized in a wide array of downstream applications within drug discovery and characterization. Furthermore, drug targets embedded in Salipro nanoparticles are functional and structurally intact, as has been demonstrated by a wide range of biochemical and biophysical methods such as cryo-electron microscopy, nuclear magnetic resonance spectroscopy, and enzymatic assays.12–16
Case study: Generation of high affinity mAbs specific to structural epitopes of an ion channel
In this work, Salipro Biotech explored the potential of the Salipro technology for mAb discovery by using a synthetic human antigen–binding fragment (Fab) phage display library, in collaboration with Helena Persson and Camilla Hofström from SciLifeLab (Stockholm, Sweden) as well as Christian Löw from the European Molecular Biology Laboratory (Hamburg, Germany). To enable efficient phage selection, the Salipro scaffold protein was biotinylated and used to reconstitute an ion channel into biotin–Salipro–ion-channel nanoparticles, as schematically illustrated in Figure 2.
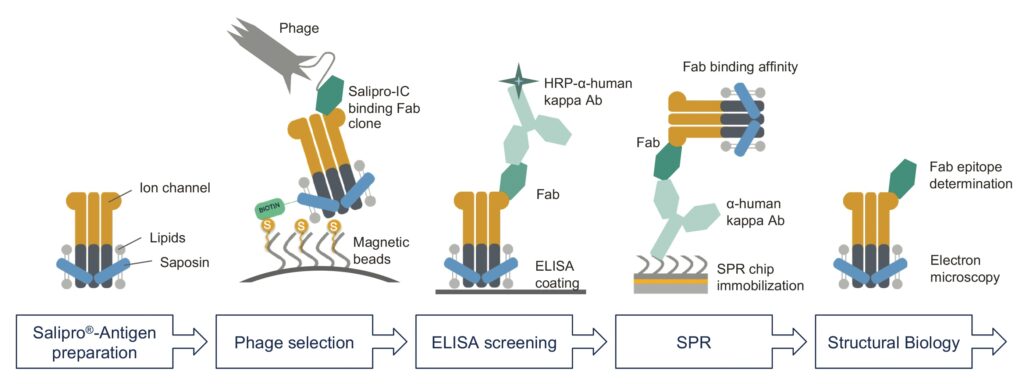
Biotin–Salipro–ion-channel particles were used as an antigen in a standard phage display selection workflow that had previously been optimized for soluble protein antigens.17 Selected Fab clones were initially screened by ELISA, and positive hits were sequenced to identify unique clones. Cleared culture media from all the unique clones containing expressed Fab were then used for an initial antigen binding test by surface plasmon resonance (SPR) spectroscopy. The seven Fab clones with the highest relative affinity were selected for protein purification and further characterization.
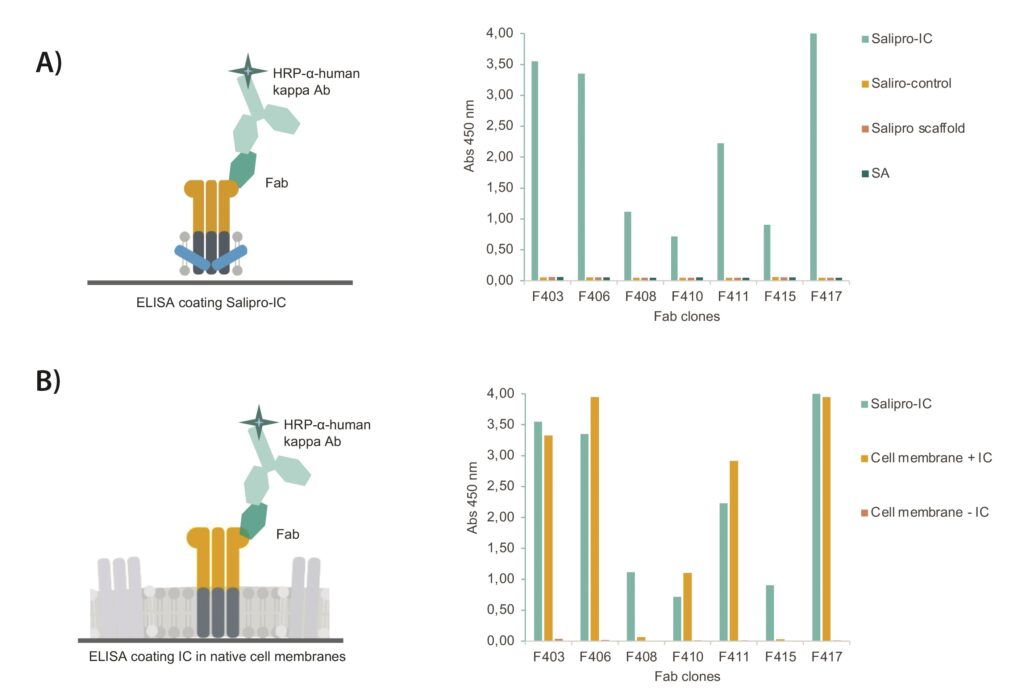
Binding specificity of purified Fabs was determined by ELISA. ELISA wells were coated with Salipro–ion-channel particles, and control wells were coated with Salipro–control particles (containing another multiple transmembrane protein), Salipro scaffold protein alone, or streptavidin (SA). All seven Fab clones showed clear, specific interactions with Salipro–ion-channel particles without any detectable unspecific interactions to any of the control proteins (Figure 3a).
Next, Fab binding to native ion channels was explored in cell membranes. Crude membranes were prepared from cells with and without the ion channel expressed, followed by an ELISA binding analysis. Five of seven Salipro–ion-channel-binding Fabs bound to epitopes present on the membrane-bound ion channel. None of the Fab clones interacted nonspecifically to membranes lacking expression of ion channel (Figure 3b).
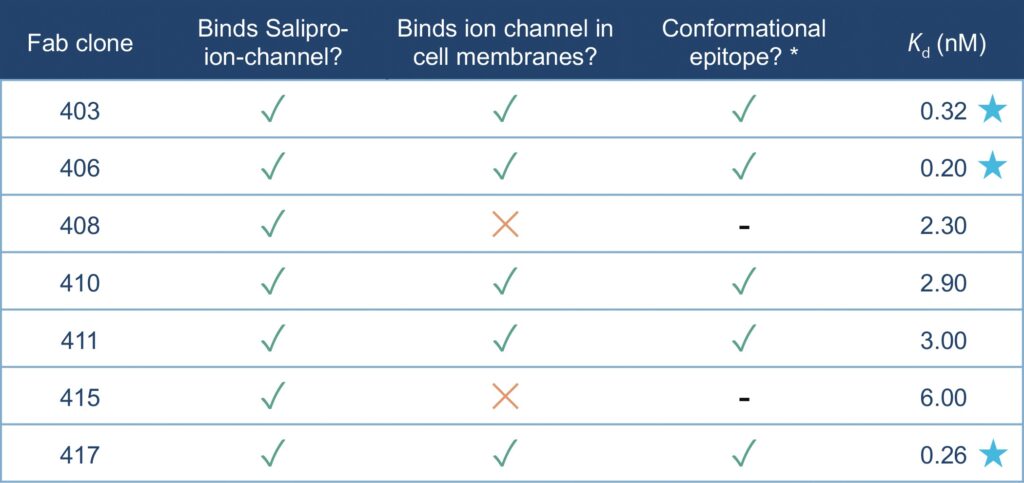
To investigate Fab binding kinetics, SPR spectroscopy was conducted using α-human kappa antibody immobilized to a CM5 chip (Cytiva). Purified Fab clones were then captured as ligands, in turn enabling binding measurements of the Salipro–ion-channel nanoparticles as injected analyte (schematically illustrated in Figure 4). Three Fab clones bound with high affinity to Salipro–ion-channel particles with binding constants (Kd values) in the 200–300 pM range (clones 403, 406, and 417). The additional four Fab clones had Kd values in the range of 2.3–6.0 nM (clones 408, 410, 411, and 415). All biophysical Fab binding data are
summarized in Table 1.

Summary
The Salipro platform technology enabled phage display selection and downstream antibody characterization with a purified multiple transmembrane ion channel. Selected high-affinity Fab clones bound to structural epitopes on native ion channels.
In conclusion, the Salipro platform technology enables a straightforward workflow to generate and characterize high-affinity binders toward GPCRs, ion channels, and transporters. Thus, the Salipro technology represents an opportunity to mine the vast untapped potential of these important classes of therapeutic targets.
Anne-Sophie Tournillon, PhD, is a scientist at Salipro Biotech. Robin Löving, PhD ([email protected]), is the company’s chief scientific officer.
