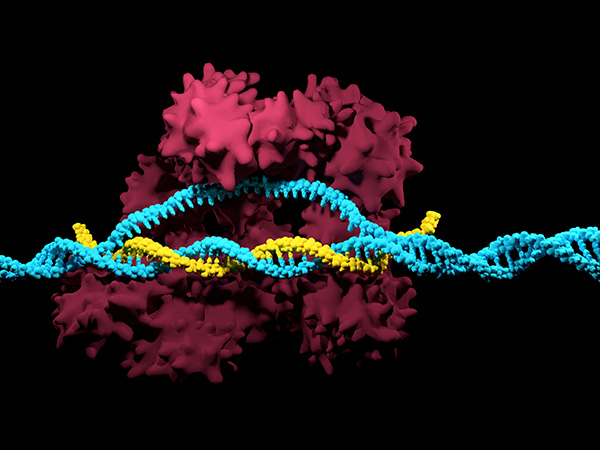
When you wield a sharp cutting tool, you need a steady hand. Otherwise, slip-ups are bound to happen, and you’ll mar your work—whether you’re performing carpentry or editing the genome. In the case of CRISPR-Cas9 genome editing, the equivalent of a steady hand is guide RNA, the RNA molecule that can, by dint of its sequence, “feel” a DNA region that has a complementary sequence. Guide RNA essentially guides the cutting tool with which it is complexed, that is, the Cas9 nuclease.
According to a new study, it turns out that the guide RNA–Cas9 complex feels its way along DNA more sensitively, or less, depending on a group of amino acid residues in Cas9 that contact the phosphate backbone of the guide RNA. In some cases, altering these residues resulted in less specific Cas9 variants, cutting tools more prone to off-target effects. But in other cases, it resulted in more specific Cas9 variants, cutting tools better suited to precision work.
The study was led by Emmanuelle Charpentier, PhD, director of the Max Planck Unit for the Science of Pathogens in Berlin. “Sometimes, Cas9 can cut DNA sequences that are very similar to the actual target, known as off-targets,” she noted.
To investigate which features of Cas9 determine the sensitivity to mismatches along the target DNA, Charpentier and colleagues performed in vitro biochemical assays and bacterial survival assays in Escherichia coli. Then they demonstrated that arginines in the Cas9 bridge helix influence guide RNA, target DNA binding, and cleavage. Essentially the arginines cluster in two groups that either increase or decrease the Cas9 sensitivity to mismatches.
Detailed findings appeared March 2 in Nature Chemical Biology, in an article titled, “Bridge helix arginines play a critical role in Cas9 sensitivity to mismatches.”
“We show that the bridge helix is essential for R-loop formation and that R63 and R66 reduce Cas9 specificity by stabilizing the R-loop in the presence of mismatches,” the article’s authors wrote. “Additionally, we identify Q768 that reduces sensitivity of Cas9 to protospacer adjacent motif-distal mismatches.”
In addition, the article emphasized that one of the researchers’ Cas9 variants, R63A/Q768A, increased the gene editing specificity of Cas9 also in human cells.
If Cas9 variants are more sensitive to mismatches, they are less likely to produce unwanted cuts—and inaccuracies in genome editing. “An unintended cut at the wrong place in the human genome can have profound consequences. That is why we need a more specific system,” stressed Michael Böttcher, PhD, a study co-author and assistant professor at the Martin Luther University Halle-Wittenberg.
Scientists are trying to optimize Cas9 specificity using different approaches. In the current study, the team of researchers from Berlin and Halle focused on an evolutionarily conserved domain of Cas9, known as bridge helix.
The scientists found that the bridge helix plays a critical role in the mechanism by which Cas9 interacts with its guide RNA and DNA target site. They identified a group of amino acid residues that contact the phosphate backbone of the guide RNA, thereby facilitating the formation of a stable loop, which is essential for the activity of Cas9. In such a loop, the Cas9-bound guide RNA pairs with the complementary strand of the DNA target sequence while displacing the second DNA strand, thereby enabling Cas9 to cut both DNA strands.
“Our results provide a new basis for further optimization of CRISPR-Cas9,” Charpentier concluded. “They demonstrate the need to gain more knowledge about the biochemistry of CRISPR-Cas systems to further improve them.”