Melanoma is the least common type of skin cancer but one of the most deadly, and the survival rate for advanced disease is just 20%. Although melanoma is curable when detected early, diagnosis is challenging, and misdiagnosis of cutaneous melanoma is among the most significant contributors to medical malpractice lawsuits in the United States.
Scientists headed by a team at the University of California, San Francisco (UCSF), now report on a new approach to diagnosing melanoma that uses microRNA (miRNA) biomarkers to indicate the presence of even just a few melanoma cells in biopsies of skin tumors that may contain predominantly benign cells.
“miRNAs are highly stable and measuring them is cheap, easy, and requires little tissue,” commented lead investigator Robert L. Judson-Torres, PhD, formerly at UCSF and currently with Huntsman Cancer Institute and the department of dermatology at the University of Utah, Salt Lake City. “Importantly, this new approach can identify just a few malignant melanoma cells, even when the majority of a tumor is benign. The early and accurate diagnosis of melanoma is paramount for optimizing patient outcome … Our hope is that with full clinical validation of our approach, all patients can receive the same level of diagnostic accuracy …”.
Judson-Torres and colleagues reported their findings in the Journal of Investigative Dermatology, in a paper titled, “MicroRNA Ratios Distinguish Melanomas from Nevi.”
Advanced melanoma causes more than 10,000 deaths in the U.S. every year, but differentiating between malignant lesions and the more common benign lesions, such as melanocytic nevi—which we know as moles—is challenging, the authors explained. The clinical standard for diagnosing tumors relies on histopathological assessment of formalin-fixed paraffin-embedded (FFPE) biopsy specimens. However, even the most experienced pathologists may not make the same diagnosis on the same specimen. “ … a considerable rate of discordance in diagnoses has been established even among expert pathologists,” with studies suggesting inter-observer discordance rates as high as 75%, and intra-observer discordance rates of up to 65%.
“Although cutaneous melanoma is curable when detected early, the process of differentiating between malignant lesions and the more prevalent benign lesions, such as melanocytic nevi, is challenging,” Judson-Torres stated. “Discordance rates across dermatopathologists and care centers are high. Consequently, whether a patient is accurately diagnosed early can depend on where the patient lives.”
The authors pointed out that molecular biomarkers could offer an alternative “robust, objective, and quantitative” measurement of disease state. One of the strengths of a molecular approach to melanoma diagnosis is that biomarkers could allow the development of a truly objective test that delivers an accurate, reproducible diagnostic score, regardless of who performs the assay.
The issue here is which biomarkers could best be used. Molecular tests based on gene expression may not be ideal as they could be easily confounded by tumor and tissue heterogeneity,” the team pointed out. In contrast, small, noncoding miRNAs exhibit tissue- and cell-specific expression patterns during development, and are misregulated in disease, so could represent potentially valuable candidate biomarkers for many disorders, including melanoma diagnosis, they suggested. The miRNA transcriptome is also relatively small and so more easily profiled than the mRNA transcriptome using next-generation sequencing, and the integrity of miRNAs is retained in FFPE samples, in contrast with longer RNA transcripts that are commonly degraded. “Thus, if the miRNA expression levels provided reliable insight into the progression state of a FFPE melanocytic neoplasm biopsy, they could assist in the diagnosis of difficult to diagnose cases where material is limiting,” the authors suggested.
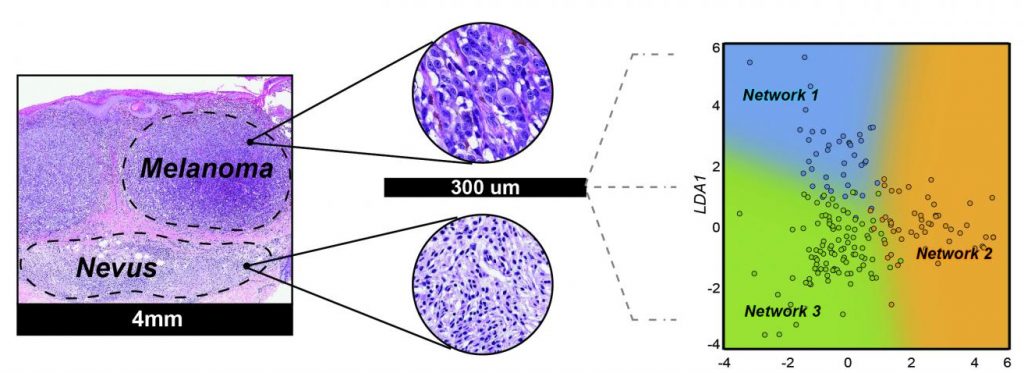
For their reported study the scientists used machine learning to develop an miRNA-based approach for consistently differentiating malignant melanoma from benign moles. They first identified tumor heterogeneity and patient age as the variables that most frequently confuse miRNA-based melanoma diagnostics. They then applied a machine-learning approach to identify miRNA expression ratios that were predictive of a melanoma diagnosis, irrespective of these key confounding variables. They tested the diagnostic relevance of their miRNA ratios on a set of validation samples comprising 82 biopsied melanocytic lesions with known outcomes (41 neoplasms diagnosed as nevi and 41 diagnosed as melanoma) from the archives of the UCSF dermatopathology section. The results indicated that the test could diagnose lesions with 81% sensitivity and 88% specificity. Importantly, the diagnoses were not influenced by tumor heterogeneity or patient age. “This performance is comparable to other molecular tests for distinguishing benign melanocytic nevi from melanomas, including chromosomal analysis by fluorescence in situ hybridization,” the authors commented.
The collective results of multiple studies have identified more than 500 miRNAs that are present at higher levels in nevi or in melanomas, but most of these have not been reproduced using independent validation sets. As part of their strategy, Judson-Torres and colleagues refined this list down to six miRNAs that could reproducibly distinguish nevi from melanoma across independent datasets and profiling platforms. “The final list of feature selected miRNAs contained two miRNAs with increased expression in melanomas (MIR31-5p, MIR21-5p) and four miRNAs (MIR211-5p, MIR125A-5p, MIR125B-5p, and MIR100-5p) with decreased expression in the melanomas,” they wrote. “Of the six miRNAs in our signature, three (MIR211-5p, MIR21-5p, and MIR125B-5p) have been previously linked to melanoma, and changes in their expression validated by in situ hybridization.”
Using these six miRNAs the investigators identified eight miRNA ratios that were predictive of diagnosis, but which were unaffected by key confounding variables. “By employing the ratios of microRNAs that were either enriched or depleted in melanoma compared to the nevi as a normalization strategy, we developed a model that classified all the available published cohorts,” the team commented. “We found that by developing a classifier based on a ratio of diagnostically important miRNA we could provide a more robust biomarker that was less susceptible to changes in tumor cell content and platform allowing for a test that could be used on a greater variety of patient samples,” said Rodrigo Torres, PhD, a postdoctoral scholar at UCSF and the study’s first author.
“In addition to improving the diagnostic accuracy for melanoma, the technique also has the potential to help catch melanomas earlier, when the tumor is entirely curable, which would significantly impact patient care,” stated study co-author Maria L. Wei, MD, PhD, who is director of the UCSF Helen Diller Family Comprehensive Cancer Center Melanoma Surveillance Clinic. The team acknowledged that further studies will be needed to confirm the accuracy of their method in ambiguous cases, and to determine whether miRNA ratios can predict metastasis or overall survival. Nevertheless, they concluded, ”Regardless of their precise functional role or their local cell of origin in the context of melanocytic neoplasia, our analyses demonstrate that the relative expression ratios of these six miRNAs can assist in distinguishing benign melanocytic nevi from malignant melanoma in FFPE samples.”

