February 15, 2011 (Vol. 31, No. 4)
Polymerase Chain Reaction Proving Its Mettle In Yet Another Arena: Clinical Tests
The polymerase chain reaction (PCR), long a tried-and-true tool in the research setting, is increasingly the basis for clinical tests aimed at improving disease diagnostic and prognostic capabilities, monitoring disease progression, and, increasingly, predicting drug response and adverse reaction or toxicity risk.
Recent PCR-based product development announcements and presentations at scientific meetings highlight the growing role this technology is playing in the translation of biological, physiological, and pharmacological information from the laboratory to the clinic.
Continuing technology improvements are contributing to the development of robust and easy-to-perform PCR-based assays that are moving toward point-of-care applications. For example, Spartan Bioscience recently received CE IVD Mark approval for its RX CYP2C19 in vitro diagnostic test, a point-of-care system designed to test a buccal swab sample for the presence of the *2 variant of the cytochrome P450 2C19 gene.
Patients carrying this gene variant—which is present in about 30% of the population—have a reduced capacity to metabolize the anticoagulant drug clopidogrel (Plavix®) into its active metabolite.
Spartan reports that carriers of the gene who receive a standard dose of Plavix following insertion of a cardiac stent to open a blocked artery have a 42% higher risk of death, stroke, or heart attack during the first year compared to noncarriers. The test can help guide drug treatment decisions.
Larry D’Andrea, CEO of Spartan, describes the Spartan RX as a “sample-to-result” testing platform that is 100% sensitive and specific. A buccal swab sample is placed directly in a tube, which is loaded into the device. The PCR-based test determines the presence of the *2 variant and can indicate whether the patient is homozygous or heterozygous at the CYP2C19 locus.
Quantifying Predictive Biomarkers
Compendia Bioscience recently introduced the first of its Tumor Segregation Panels™—the Breast Cancer Segregation Panel™ Assay—in partnership with Althea Diagnostics (www.altheadx.com). The companies also plan to launch Colon Segregation Panel™ and Lung Segregation Panel™ assays later in 2011.
The breast cancer assay uses quantitative RT-PCR to measure the expression of a panel of 96 genes selected to represent key areas of molecular variability in the breast cancer genome. The test is performed on FFPE tissue samples. Analytical algorithms used to evaluate the results can identify correlations between gene-expression variability in individual breast tumors and clinical endpoints such as drug response. The goal is to translate this information into predictive biomarkers that can form the basis for companion diagnostics for standard-of-care therapies and new, more targeted drugs in development.
In designing the Tumor Segregation Panels, Compendia’s aim is “to build a multipurpose tool that measures all the core modules [gene-expression variables for a particular tumor type] and has a clear path to market as a companion diagnostic,” says Daniel Rhodes, Ph.D., CEO and co-founder of Compendia.
These “core modules” provide comprehensive coverage of the complexity and variability of breast cancer, according to Dr. Rhodes, who believes the company’s RT-PCR assays can replace custom gene signatures used to predict parameters such as prognosis and drug efficacy. The assays yield quantitative results that can be used to define, more precisely, characteristics such as tumor grade, proliferative risk, and drug response, and to quantify biomarkers such as estrogen receptor or Her2 positivity. Dr. Rhodes will describe the technology underlying the tumor segregation panels at the upcoming “Molecular Medicine TriConference.”
The 25 core gene modules selected for Compendia’s breast cancer panel represent patterns of genes that showed variation across a cohort of more than 5,000 breast tumors. Each module is represented in the assay by two to three marker genes. The 96-gene panel can be assayed against four samples simultaneously. Althea performs the test in its CLIA laboratory on FFPE biopsy or surgical specimens collected during a clinical trial. Compendia and Althea then analyze the results and correlate the data for individual modules or combinations of modules depending on the clinical endpoints defined in the trial.
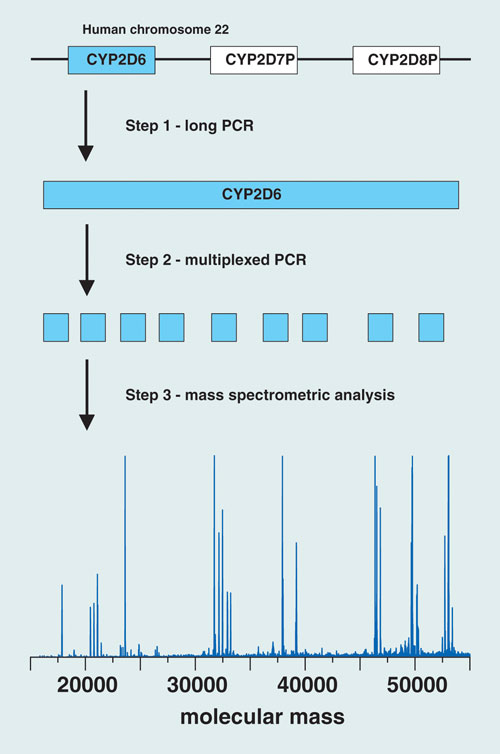
At the “San Antonio Breast Cancer Symposium” late last year, Beate Beer and colleagues from Innsbruck Medical University presented work focused on the cytochrome P450 enzyme designated as 2D6, which has a role in metabolizing about 25% of the prescription drugs presently on the market.
Tamoxifen, an anti-estrogen drug used to treat breast cancer, is one example of a drug metabolically activated by CYP2D6. Risk of adverse drug reactions or of absent or enhanced drug activity may affect patient populations that have variations in the CYP2D6 gene resulting in a nonfunctional, slow-acting, or super-fast acting form of the enzyme. Eighty CYP2D6 alleles have been identified.
The Austrian group of researchers has developed an assay strategy for CYP2D6 genotyping that combines PCR, ion-pair reversed-phase HPLC, and electrospray ionization time-of-flight mass spectrometry. Their goal is to validate the clinical relevance of CYP2D6 genotyping and its use in tailoring more effective, personalized treatment regimens to combat breast cancer.
The PCR component of the assay isolates the gene of interest and amplifies targeted regions in the CYP2D6 gene using a two-step protocol. The first step generates a long-PCR copy of the entire gene. An aliquot of the long-PCR copy then serves as a template for the second PCR step, a 20 µL multiplexed reaction that amplifies nine variable regions of the gene, which cover 604 nucleotide positions (approximately 12% of the entire gene sequence) and 27 known polymorphic sites, enabling the resolution of at least 37 known CYP2D6 alleles. HPLC-MS analysis of the PCR amplicons yields molecular mass information that serves as a sensitive measure of the nucleotide composition of each amplicon.
“By determining the nucleotide composition of an amplicon, nucleotide variations occurring within the amplified region can be detected”—including unknown variations in vicinity to the polymorphic site of interest, “which enables the resolution of subvariants of common alleles,” says co-author Herbert Oberacher.
The researchers described the use of their trimodal strategy for CYP2D6 genotyping in 199 individuals: 106 patients with breast cancer who were taking tamoxifen, and 93 healthy subjects. The method yielded “successful amplification of DNA isolated from blood and buccal swabs,” says Oberacher.
They identified 14 CYP2D6 alleles, forming 39 different genotypes, and used the information to categorize the samples according to predicted enzyme activity. Statistical evaluation of the observed frequencies revealed no significant differences between patients and healthy individuals.
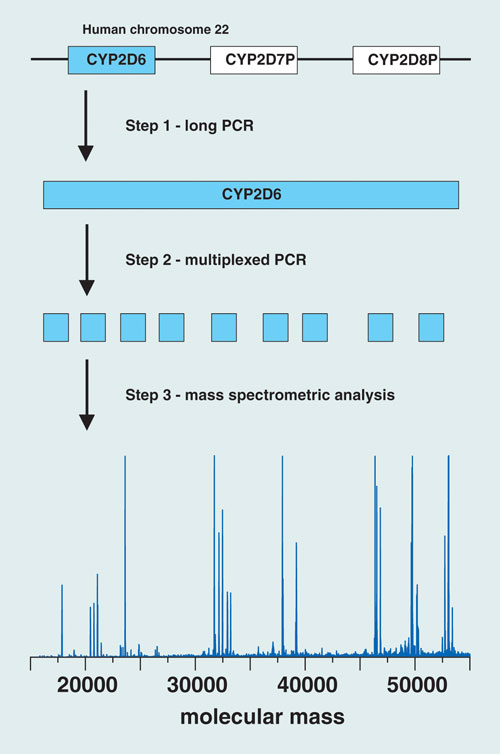
Researchers at the Innsbruck Medical University have developed an assay strategy for CYP2D6 genotyping that combines PCR, ion-pair reversed-phase HPLC, and electrospray ionization TOF mass spec.
Predicting Drug Efficacy
France Carrier, Ph.D., associate professor of radiation oncology, Marlene and Stewart Greenebaum Cancer Center, University of Maryland, envisions an opportunity to use PCR as the basis for a prognostic tool to predict a patient’s responsiveness to anticancer drugs that inhibit DNA synthesis and, thereby, cell replication. Dr. Carrier discussed his research at CHI’s recent “Biomarker Assays Development” conference.
Specifically, Dr. Carrier has developed a patent-pending technique to measure breaks in genomic DNA that result from anticancer drugs that interfere with DNA synthesis, such as topoisomerase inhibitors. Topoisomerases are enzymes that facilitate DNA replication by creating and then religating nicks in the DNA to alleviate torsional stress as the strands unwind. They preferentially cut DNA at particular sites. Treatment with a topoisomerase inhibitor that compromises the enzyme’s function will result in more DNA breaks, which prevent DNA polymerase from reading and copying the affected strand.
Dr. Carrier points to combining RT-PCR with conventional Stop-PCR as the enabling technological advance for this predictive assay strategy. It provided the sensitivity needed to be able to quantify the level of DNA damage produced by typical therapeutic doses of topoisomerase inhibitors.
Using RT-PCR to amplify genomic DNA from cells exposed to a topoisomerase inhibitor, Dr. Carrier can assess the efficiency of the drug. The more efficient it is, the less amplification there will be of the DNA sequences it targets. Based on the predictive power of this inverse correlation, it may be possible to evaluate in vitro the potential effectiveness of a drug that interferes with DNA synthesis, enabling a more personalized approach to the selection of anticancer drugs and combination therapies.
Dr. Carrier has used the assay to study peripheral blood mononuclear cells (PBMCs) isolated from the blood of treated patients and has demonstrated its utility at therapeutic drug doses. The next step toward developing a clinically applicable predictive tool will be to conduct animal studies designed to compare conventional measures of drug efficacy (tumor shrinkage) with PCR-based quantification of DNA breaks in cells derived from biopsy samples and PBMCs.
Powerful Prognostic Tools
Dave Hoon, Ph.D., director of molecular oncology at the John Wayne Cancer Institute, will also speak at the “Molecular Medicine TriConference”. He will present the results of two Phase III multicenter randomized clinical trials that demonstrate that circulating tumor cells (CTCs) are an independent prognostic factor for disease-free and overall survival in patients with stage 3 and 4 melanoma. Dr. Hoon and colleagues developed a multimarker real-time quantitative reverse transcriptase assay designed to detect and quantify CTCs in the blood of patients with melanoma.
The RT-qPCR assay measures the levels of four melanoma-associated CTC mRNAs: MART-1, GalNAc-T, PAX3, and MAGE-A3. In Phase II trials, the presence of and increasing numbers of these biomarkers, measured serially in patients undergoing combination immunotherapy and chemotherapy, correlated with disease progression, treatment response, and overall and disease-free survival.
“This is a direct blood assay” performed on mRNA isolated from whole cells in patients’ blood samples, says Dr. Hoon. Its advantages include being real-time, probe-based, highly specific, and optimized for the biomarkers selected. The rationale behind designing a multimarker assay was based on the heterogeneity of tumors and CTCs, making it unlikely that a single marker would be predictive, particularly in aggressive disease with different sites of origin.
“Tumor cells that express these markers are more aggressive. It is not the number of CTCs that is prognostic; rather it is the content of the cells,” Dr. Hoon says. The goal is to develop this assay as a blood test for use in predicting which treatment is likely to be most effective following surgery, for determining disease spread, and for monitoring the effectiveness of adjuvant therapy.
New DNA Polymerase from New England Biolabs
PCR applications can benefit from the new OneTaq DNA Polymerase just introduced by New England Biolabs, which amplifies routine amplicons as well as AT-rich and GC-rich templates, according to company officials.
“OneTaq enables scientists to use one polymerase to achieve high yields and specificity for all templates, regardless of GC content,” states Fiona Stewart, product marketing manager for PCR polymerases. “For amplifications requiring a hot start modification, OneTaq Hot Start provides additional convenience by requiring no activation incubation and allows room temperature reaction set-up.”
OneTaq combines Taq and Deep Vent® DNA polymerases. It is available in two formats: Standard and Hot Start. The Hot Start version includes an aptamer-based inhibitor that binds reversibly to block polymerase activity at temperatures below 45°C. The OneTaq product includes two reaction buffers, standard and GC. A High GC Enhancer solution can be added to the GC buffer mix for amplifying templates with very high GC content.