May 15, 2008 (Vol. 28, No. 10)
SciGene’s Hybex Incubation System Facilitates More Uniform Hybridization
Cytogenetic analysis of chromosomes provides the foundation for the clinical diagnosis of genetic disorders caused by chromosomal abnormalities. Initially, chromosome banding techniques such as G-banding were used as diagnostic tools. Banding techniques, however, lack the specificity and resolution needed for the diagnosis of a wide variety of chromosomal abnormalities.
Molecular cytogenetics (fluorescent in situ hybridization or FISH) combines classical cytogenetics with molecular techniques, which significantly increases the resolution and specificity of cytogenetic analysis. FISH with chromosome-specific probes reveal hybridization signals, even on inadequately spread metaphase chromosomes and interphase nuclei. It is capable of detecting subtle chromosomal abnormalities that are not detected using conventional cytogenetic analysis. Additionally, FISH allows quantitative analysis of target sequences, thus providing an advanced tool for genetic research.
During FISH, a fluorescent-labeled nucleic acid probe with sequences complementary to the area of interest on a chromosome is hybridized to chromosome spreads or nuclei. Probes bound to the target sequences are visualized by fluorescence microscopy.
During multicolor FISH, the use of fluorescent dyes with different emission wavelengths allows for the use of multiple probes during a single hybridization to simultaneously target different chromosomes or different areas within a chromosome. FISH can be qualitative (to study the presence, absence, or position of a sequence) or quantitative (e.g., to study the length of a repeat sequence).
In studies requiring quantitative measurement of a target sequence such as telomere-length measurement, it is critical to have slide-to-slide and day-to-day reproducibility to enable data comparability across various hybridizations. It is also crucial to have uniform hybridization to minimize the signal variations within a slide.
Current methodologies often utilize various sealants around a cover slip to create a sealed chamber, and hybridizations are incubated in a conventional oven. These methods generate data with high day-to-day variability in fluorescence intensity and are susceptible to photo-bleaching of fluorescent dyes.
In this article, we describe a new method for performing quantitative telomere FISH using the Hybex® microarray incubation system from SciGene (www.scigene.com). The Hybex hybridization chambers have advanced sealing to prevent loss of humidity and photo-bleaching, resulting in a more uniform hybridization and brighter signal, which are critical for quantitative FISH that requires high sensitivity and reproducibility. We have applied this procedure for molecular epidemiological studies and obtained highly reproducible results.

Figure 1
Slide Preparation
Metaphase chromosome spreads were prepared following standard cytogenetic procedures. Lymphocyte cultures were established by inoculating 1 mL of fresh blood into 9 mL of blood culture medium (RPMI-1640 medium supplemented with 15% bovine serum and 1.5% phytohemagglutinin). The cultures were incubated at 37ºC for four days. To arrest the cells at metaphase, 0.2 µg/mL colcemid was added to the culture one hour before harvest. The cells were treated in hypotonic solution (0.06 M KCl) at RT for 25 minutes.
The chromosome preparation from cultured lymphocytes were dropped onto a clean glass slide, fixed in 3:1 methanol:acetic acid for one hour at room temperature, and then air-dried. Slides were then dehydrated in an ethanol series (2 minutes each in 70%, 80%, 90%, and 100% ethanol) and then air-dried for 15 minutes.
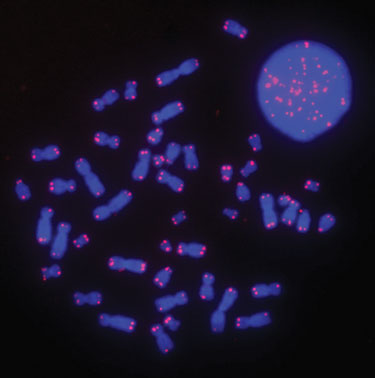
Figure 2
Denaturation and Hybridization
Fifteen microliters of hybridization mixture consisting of 0.3 µg/mL Cy3-labeled, telomere-specific PNA probe; 0.1 µg/mL FITC-labeled, chromosome 9-specific Sat III PNA probe; 50% formamide; 10 mM Tris-HCl, pH 7.5; 5% blocking reagent; and 1X Denhart’s solution was applied to each slide and covered under a cover slip.
Before starting the procedure, SciGene’s Hybex microarray incubation system was preheated to 75°C with a water bath insert in position. Fifteen microliters of hybridization mixture was pipetted onto each slide, and the spread area was covered with a cover slip, avoiding the creation of air bubbles.
Then, the slides were inserted into the Hybex slide racks, which were mounted on a hybridization chamber base (Figure 1A). One milliliter of water was pipetted on the absorbent pad situated in the chamber cover to provide adequate moisture during hybridization. The cover of the chamber was sealed to the base by tightening the corner screws (Figure 1B), and a black handle attached. The water bath insert was removed from the preheated Hybex unit, and the assembled chamber was placed into the heating unit, using both chambers to ensure accurate heat control (Figure 1C).
Once the system temperature equilibrated to 75°C (in 8–10 minutes), the slides were incubated for five minutes. Afterward, the hybridization chambers were removed and left at room temperature for 15 minutes. The Hybex unit was speed-cooled by setting the temperature to 30°C and placing a water bath insert into the unit. When 30°C was reached, the hybridization chambers were placed back into the instrument and incubated for three hours. (For DNA probes, hybridization can be performed at 37–42°C for 10 to 16 hours.)
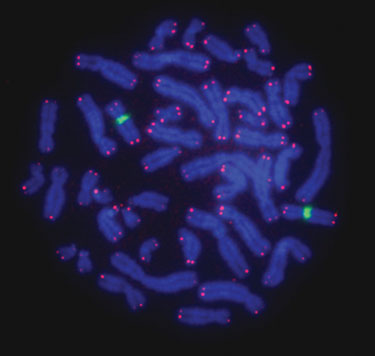
Figure 3
Posthybridization Processing of Slides
Four staining dishes were prewarmed in a water bath set at 42°C, each contained 200 mL of 2X SSC, 1X SSC, 0.5X SSC, and 0.1X SSC. The hybridization chambers were removed from the Hybex incubator, corner screws loosened, covers lifted off, and slide racks removed. The racks were placed into the dish containing 2X SSC at 42°C for five minutes. The processing handle was attached to the slide rack and agitated up and down to release cover slips.
The slide rack was then moved to a second dish containing 1X SSC at 42°C and soaked for 10 minutes. The wash was repeated with 0.5X SSC and 0.1X SSC for ten minutes each at 42°C. Slides were dehydrated in the ethanol series (two minutes each in 70%, 80%, 90%, and 100%) and air-dried. Following the drying of slides, 15 µl of antifade solution with DAPI was added to each slide, and spread area was covered with a cover slip.
Slide Visualization
The sample slide was analyzed using a Leica DM 4000 epifluorescence microscope equipped with short-arc mercury lamp illumination, a 100X/1.3 N.A. oil immersion neofluotar objective lens, and appropriate band pass filters for Cy3, FITC, and DAPI. Fluorescent images were captured with a charge-coupled device camera.
Results
Figures 2 and 3 demonstrate that the FISH procedure using the Hybex microarray incubation system can be utilized to generate bright and high-quality telomere signals. We also observed that the brightness of telomere signals was uniform across different areas of the slide with low background noise. The improved signal-to-noise ratio increased the sensitivity of the method, allowing for the measurement of short telomeres in cancer cells (data not shown).
To test the reproducibility of the method, we performed three repeat experiments one week apart, each with five samples. The Table shows that the coefficient of variation of these repeat experiments is 9%. The method described in this article shows significant improvement over the method previously used in our laboratory with a conventional hybridization oven, delivering data with high reproducibility.
Rajiv Raja, Ph.D. ([email protected]), is director of genomics applications at SciGene, Zuguo Li, Ph.D., is postdoctoral fellow, and Yun-Ling Zheng, Ph.D., is assistant professor at Lombardi Comprehensive Cancer Center at Georgetown University.
Web: www.scigene.com



