September 1, 2012 (Vol. 32, No. 15)
Bee Na Lee Ph.D.
Changing Binding Conditions Allows for Recovery from Cell Culture, FFPE, and Tissue Samples
There is great interest in understanding how miRNA can be used as a biomarker for cancer development and cancer prognosis, and discovering how it might be used as a therapeutic target for cancer treatment.
With high demand for, and the fast growth of, miRNA research, an efficient and robust miRNA purification method was needed to enhance downstream assays. Most nucleic acid extraction procedures do not favor miRNA recovery, as smaller nucleic acids such as miRNA are removed during purification processes. In this article, two methodologies for recovering miRNA using the Agencourt SPRI chemistry (Beckman Coulter Life Sciences) are described.
The solid-phase reverse immobilization (SPRI) procedure is an easy, rapid, high-yield, and automation-friendly miRNA isolation method that does not require organic solvents, centrifugation, or filtration steps. This method uses carboxyl-coated magnetic particles that reversibly bind nucleic acid in the presence of binding buffers and crowding reagents.
Typically, there are three basic steps in the purification procedure (Figure 1). In the first step, nucleic acid immobilization, SPRI beads are directly added to sample reactions. Nucleic acids are immobilized onto the SPRI beads, leaving contaminants in solution. In the second step, contaminant removal and wash, a magnetic field is used to pull the microparticles out of solution. Contaminants are aspirated, and microparticles are thoroughly washed with molecular biology-grade ethanol, yielding high-quality nucleic acids. In the third step, purified nucleic acids are easily eluted from the microparticles under aqueous conditions, which provides maximum flexibility for downstream applications.
Agencourt SPRI chemistry was designed to selectively capture larger nucleic acids; any fragments smaller than 50 nucleotides are usually removed during the binding steps. miRNAs have an approximate size of 22 nucleotides: Our initial test for miRNA isolation using the Agencourt RNAdvance Cell v2 protocol monitored the recovery of the 22 nt oligonucleotides through various binding and rebinding buffer conditions.
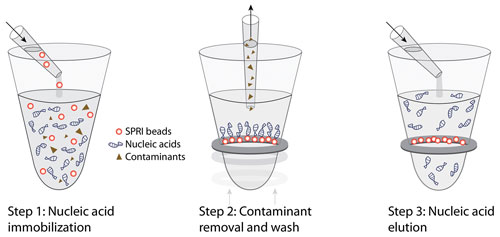
Figure 1. Solid-phase reverse immobilization (SPRI) technology requires three steps.
The results showed that recovery of the 22 nt oligonucleotides can be enhanced by changing the binding conditions in the binding and rebinding steps. For each test condition, 1 µg of oligonucleotide was used as the input sample and eluted in 40 µL of nuclease-free water, with an estimated yield of 25 ng/µL of oligonucleotide if the recovery rate reaches 100%. The recovery of the 22 nt nucleotides was then measured by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, data not shown) and ethidium bromide staining using E-Gel agarose electrophoresis (Life Technologies) (Figures 2A and 2B).
The highest yield of the 22 nt oligonucleotides (lane 9) was selected for a HeLa cell miRNA purification using the Agencourt RNAdvance Cell v2 protocol.
A similar principle was applied for miRNA isolation from the FFPE samples using the Agencourt FormaPure kit protocol. TaqMan qPCR expression assay (Life Technologies) of let-7c demonstrated that miRNA was successfully extracted from HeLa cells and from FFPE samples. It was further demonstrated that the SPRI manual extraction procedure performs comparably to the SPRI procedure performed using a Beckman Coulter Life Sciences’ Biomek FXP platform running an automated extraction method (Figure 2A).
For the cell culture experiment, frozen HeLa cells (50,000 cells per microplate well) were thawed at room temperature for 10–15 minutes. A 63 µL lysis proteinase K solution was added to each well, and the plate was incubated for 30 minutes. The cell lysate was transferred into the wells of a microplate (Abgene) containing 330 µL of freshly made binding buffer solution (80 µL Bind Buffer; Beckman Coulter Life Sciences) and 250 µL of isopropanol (American Bioanalytical).
Cell lysate was incubated with binding solution for 5 minutes, and the microplate was placed on the Agencourt SPRIPlate 96 Ring Super Magnet plate (Beckman Coulter Life Sciences) for 5 minutes to separate the beads. Supernatant was aspirated and removed from the microplate wells. The beads were washed once with 300 µL of fresh 85% ethanol (American Bioanalytical), the microplate was placed back on the magnetic plate, and the ethanol was aspirated and removed.
Sample plates were allowed to air dry on the magnetic plate for 10 minutes, miRNA was eluted with 40 µL of nuclease-free water, and the sample plate was placed back on the magnetic plate for 2 minutes. Eluted nucleic acids were transferred into a new plate. Steps for DNase treatment and rebinding can be introduced before the elution to remove genomic DNA.
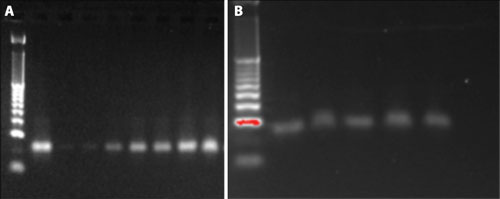
Figure 2A. Yields of the 22 nt oligonucleotides using different binding conditions: 1 µL of sample was loaded in each lane. Lane 1 was the 10 bp DNA size marker ladder, lane 2 was the control sample (25 ng of the 22 nt oligonucleotides), lanes 3–9 were loaded with 1 µL of the 22 nt oligonucleotides that were recovered from different binding conditions, with lane 9 having the highest amount of isopropanol in the binding and rebinding steps. Figure 2B. The yield from the manual extraction method is compatible with the automated extraction method using the 22 nt oligonucleotides. 1 µL of sample was loaded in each lane. Lane 1 was the 10 bp DNA size marker ladder, lane 2 was the control sample (25 ng of the 22 nt oligonucleotides), lanes 3–4 were DNA recovered from the manually extracted method, and lanes 5–6 were DNA recovered from the Biomek extraction method.
For the FFPE miRNA isolation, lysate from the FFPE samples was prepared according to the Agencourt FormaPure protocol. 200 µL of the FFPE lysate was transferred into a 96-Well Riplate–2.2 mL (WorldWide Medical Products), 150 µL of Bind 1 buffer and 20 µL of Bind 2 buffer (Beckman Coulter Life Sciences) plus 800 µL of isopropanol (American Bioanalytical) were added into the lysate, mixed by aspirating and dispensing with a pipette five times, and incubated at 55°C for 5 minutes.
The microplate was placed onto the magnetic plate for 5 minutes, the supernatant was slowly aspirated, the sample plate was removed from the magnetic plate, and the beads were washed with 750 µL of freshly made 85% ethanol by pipette mixing five times. Beads were settled on the magnetic plate again for another 5 minutes. Clear supernatant was carefully removed. The sample plate was air dried for about 10 minutes, and the miRNA and total nucleic acid were recovered with 40 µL of nuclease-free water.
Total RNA quality was measured using the Bioanalyzer 2100 (Agilent Technologies, data not shown) and the concentration of the nucleic acids was measured by NanoDrop 2000 spectrophotometer (data not shown).
Either 50 ng or 20 ng of extracted RNA was used for the TaqMan micro-RNA qPCR assay (Life Technologies). RT and PCR reactions were performed according to the manufacturer’s protocols with the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies) and TaqMan Universal Master Mix (Life Technologies).
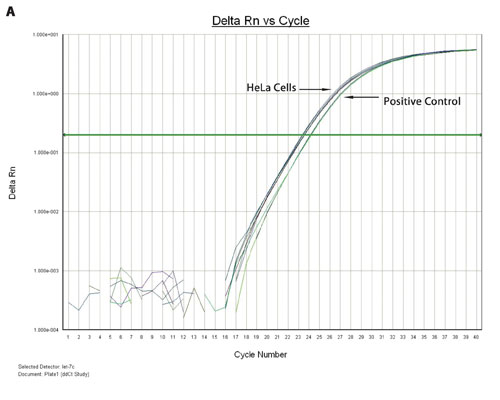
Figure 3A. TaqMan qPCR profiling of the let-7c expression in HeLa cells and FFPE extract: miRNA from a 96-well HeLa cell culture was extracted using the Biomek multichannel method, whereas the miRNA from 96-deep-well FFPE samples was extracted using the Biomek Span8 method. The amplification plots of the let 7c miRNA expression from HeLa cells and a positive control are shown.
Figures 3A and 3B show the overlaid amplification plots of the let-7c in HeLa cells, FFPE and positive control samples. The minus RT reaction and nontemplate control samples showed negative amplification, suggesting that the amplification resulted from the miRNA (data not shown).
In summary, this study showed that changing the binding conditions in the binding and rebinding steps is sufficient to recover the smaller nucleotides and miRNA from cell culture, FFPE, and tissue samples. Yield from the manual extraction method is compatible with the automated extraction method. The procedure can be accomplished in less than 90 minutes on a Biomek FXP platform with automated method for 96 samples.
Furthermore, recovery of the miRNA and total RNA can be achieved in a single process using the Agencourt SPRI chemistry. Therefore, researchers have the capability to compare the miRNA and gene expression profiles within the same sample, which allows them to save time and expense for sample preparation.
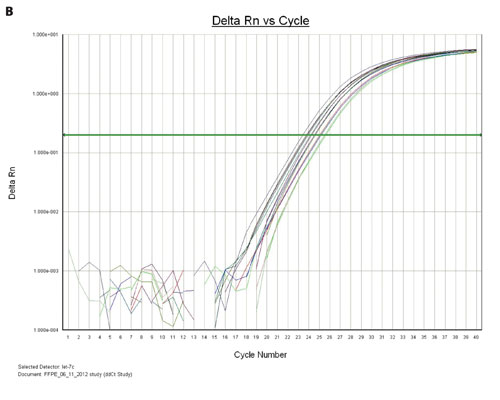
Figure 3B. The amplification plots of the let-7c miRNA from the multiple FFPE samples that were eluted with either 40 µL or 80 µL of nuclease-free water are shown.
Bee Na Lee, Ph.D. ([email protected]), is a senior application scientist with Beckman Coulter Life Sciences (sample prep team). The author is grateful to Isabel Gautreau, staff application scientist, technical support, Beckman Coulter Life Sciences, for technical advice and FFPE samples, and to Janice Linder, senior development scientist, cell culture department, Beckman Coulter Life Sciences, for HeLa cell cultures. Beckman Coulter, the stylized logo, Agencourt, Biomek, RNAdvance, SPRIPlate, FormaPure and SPRI are registered trademarks of Beckman Coulter and are registered in the USPTO. TaqMan qPCR is a registered trademark of Roche Molecular Systems.