February 15, 2009 (Vol. 29, No. 4)
Reverse Engineering/Forward Simulation Platform Reveals How Biomolecules Interact
We know the numbers by heart: bringing a new drug to market requires, on average, 12–15 years and about $1 billion. And despite the record investment in R&D in 2007 ($58.8 billion), the development of new chemical entities (NCEs) was at an all-time low. In 2007, the FDA approved just 19 new medicines—the fewest in more than two decades. Adding even further pressure to the industry are the blockbuster drugs that have been removed from the market in recent years after real-world experience highlighted safety concerns.
The growing investment in R&D combined with declining output is further reinforcing the need for change in the way drugs are discovered and developed. In an effort to improve and accelerate the process, new ways of thinking and new strategies are constantly being sought and applied.
As we entered the 21st century, biology was rapidly evolving from its classic reductionist approach, wherein a single particular gene or pathway is investigated in a limited or isolated context. We witnessed the emergence of systems biology, which offered a radical new way of thinking, focused not on individual genes and proteins but on the dynamic, integrated interactions of many biological components.
The flood of data that emerged from the human genome project and the explosion of microarray and proteomics research fueled the systems biology fire. Systems biology departments and institutes sprung up across the world. In order to make sense of the massive amounts of omics data (e.g., genomic, proteomic, metabolomic) being generated, scientists built in silico technologies and bioinformatics tools to model pathways within an increasingly complex biological landscape. Systems biology brought with it high expectations that an understanding of these complex interactions would help make predictive, preventive, and personalized medicine a reality.
The expectation was that once these models were defined, they could be used to discover the mechanisms of disease as well as drug efficacy and toxicity. Scientists and clinicians would be able to stratify diagnoses based on molecular markers, aggregate diseases previously thought to be unrelated by revealing shared biology, and gain a more comprehensive, systems-centric view of the dynamic, integrated interactions of all our genes and proteins.
These models could also be used to develop hypotheses about how a biological system will respond when perturbed, offering important insights to accelerate the drug discovery and development process. For example, the ability to predict how all the elements of a biological pathway will react to introduction of a small molecule drug can facilitate target identification and help reveal possible side effects.
Roadblocks Encountered
Key challenges were encountered during the emergence of systems biology. First, while genome sequencing and DNA microarray technologies were readily available, the cost at the time was still extremely high, thus limiting the amount of quality data being produced to build and validate these models. Second, there was insufficient computer processing speed available to effectively churn through all the omics data being produced in order to construct the system.
Over time, technology advancements allowed for dramatically faster, less expensive DNA sequencing. In parallel, advances in supercomputing began to offer researchers the processing capacity they needed to begin extracting knowledge from the crush of data in a timely manner.
But major stumbling blocks toward a holistic view of the system remained. Newly developed mechanistic models were being applied to data. While these models were able to put some of the data into context, the approach was (and remains) limited because these models are based on known biology—perhaps at most 5–10% of the potentially billions of interactions among our roughly 25,000 genes and hundreds of thousands of proteins.
The ability of these models to create meaningful simulations and enable new discoveries was further limited by their reliance on literature-based knowledge, not raw data, which inherently biases their results. The use of incomplete models to assess data limits the ability to create meaningful simulations and make accurate predictions.
Furthermore, the actual development of these mechanistic models also poses a significant challenge to researchers. Model development is a painstakingly slow, manual process, often requiring months and years.
A data-driven, reverse-engineering approach to modeling holds significant promise for bringing greater predictive power to systems biology and drug development. Starting with raw data, this new approach enables the creation of unbiased in silico “blueprints” of disease states and normal human biology that allow researchers to understand the driving perturbations underlying disease and ways of countering those perturbations.
Gene Network Sciences (GNS) has developed a Reverse Engineering/Forward Simulation platform (REFS™, Figure) that rapidly conducts billions of calculations on raw biological data (genetic and epigenetic, molecular profiling, and clinical) to reveal how the biomolecules interact with one another in the complete biological system.
Using IBM Blue Gene supercomputers with more than 30,000 processors, the REFS platform produces a blueprint that defines the most likely causal network connections among all of the variables measured and best describes the underlying system that gives rise to the raw biological data.
In a recent project with Johnson & Johnson, the REFS platform identified seven new genes that were expected to play a role in the efficacy of a cancer drug in development. siRNA knockdown experiments subsequently validated six of the seven predictions, making these biomarkers key to stratifying patient responders from nonresponders.
REFS-based models can also be queried through in silico experiments to discover the highest impact molecular targets for the disease being studied, markers related to specific drug treatments, and the effects of drug dosages or combinations at different time points. Such results, obtained in hours or days rather than months or years, can be validated easily at the lab bench.
Another application of the REFS platform is the evaluation of SNP and gene-expression data in conjunction with Phase II clinical outcomes data for the purpose of reverse engineering a model of disease in patient cohorts. REFS-based simulations can be used to identify key molecules responsible for nonresponse in cohorts. These markers can then be used to stratify patient populations for Phase III trials.
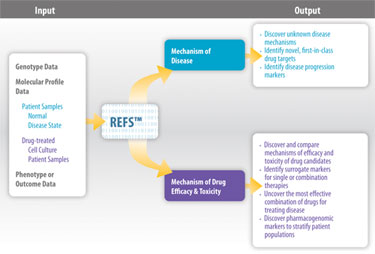
Reverse Engineering/Forward Simulation (REFS) systematically turns multiple layers of biologically relevant data into a useful view of human biology and disease as a whole.
Drug Development
Systems biology is a continually evolving field, and one that holds great promise for drug discovery and development. The ability to process and contextualize large data sets positions systems biology as a foundational element in the emerging big-data era of biomedical science. A complete, data-driven in silico view of what constitutes a disease at the molecular level could highlight the causative components of disease mechanisms, give insight into the systemic effects of a drug candidate, and give developers crucial information for making go/no-go decisions.
In 2008, Merck published two studies describing genetic susceptibility to obesity that involves changes in entire networks of genes rather than changes in just a few genes. The research also demonstrated the use of genomic techniques to understand complex changes that underlie common diseases.
Systems biology can also bring greater personalization to healthcare. Personalized medicine can only work, however, if a disease’s causative molecular alterations can be identified and counteracted. Recently published studies continue to shed light on the heterogeneity of cancer mutations, for example, and reinforce that a more thorough understanding of the molecular landscape is necessary to guide more personalized treatment.
Understanding the complete effects of a drug or drug combination in a given cell system or disease state can provide developers with the opportunity to fine-tune new candidates to maximize efficacy and reduce toxicity, or highlight new indications for existing drugs based on shared pathways or targets.
Conclusion
Systems biology is evolving to meet the needs of researchers and drug developers who must extract meaningful, applicable knowledge out of an overwhelming—and growing—amount of biological data. Armed with a greater understanding of the complexity of disease, industry will be better positioned to populate development pipelines with more promising drug candidates and help make predictive, preventive, and personalized medicine a reality.
Colin Hill ([email protected]) is CEO of Gene Network Sciences.
Web: www.gnsbiotech.com.



