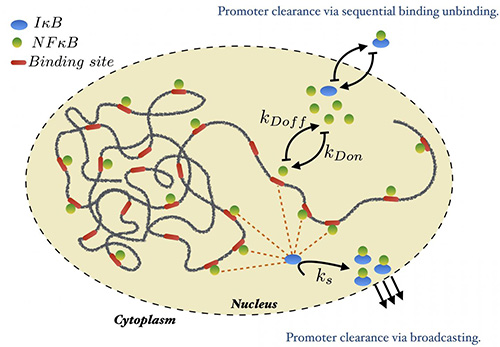
A molecular stripping mechanism is responsible for “broadcasting” genetic transcription instructions through a genetic switch known as NFkB-DNA-IkB. When triggered, the IkB inhibitor (blue) tells NFkB proteins (green) to release from DNA binding sites (red) to stop transcription of multiple genes at once. The IkB protein binds to NFkB and forces a physical twist in the protein that releases its grip on DNA. [Wolynes Lab/Rice University]
The regulatory elements known as genetic switches are usually understood in terms of thermodynamics. But thermodynamics, as a way of explaining physical processes, gives us just half the picture—and not necessarily the half that interests us. The other half is about kinetics. Essentially, thermodynamics is all about the “if” of a process, whether or not it will occur. Kinetics is all about the “how,” the factors that account for the duration of a process.
This dichotomy, familiar to all students of chemistry, is being revisited by researchers who hope to give us a full picture of gene regulation—not just the thermodynamic part, which is all about binding affinities, but also the kinetic part. In the case of a particular gene switch, the kinetic part involves the opposite of binding. It’s about letting go.
At Rice University, a team of researchers led by theoretical biological physicist Peter Wolynes, Ph.D., has found that a class of gene switches can be understood best in terms of kinetics, not thermodynamics. These gene switches are based on the NF-κB/IκB/DNA system and are master regulators of an array of cellular responses.
According to Dr. Wolynes’ team, recent kinetic experiments have shown that IκB can actively remove NF-κB bound to its genetic sites via a process called “molecular stripping.” This allows the NF-κB/IκB/DNA switch to function kinetically rather than thermodynamically, countering decades-old classical models in molecular biology that attribute the control of genetic processes to the thermodynamics of biochemical reactions.
The team described its surprising findings December 23 in the Proceedings of the National Academy of Sciences (PNAS), in an article entitled, “Molecular stripping in the NF-κB/IκB/DNA genetic regulatory network.” This article suggests that any delays in unbinding from the numerous NF-κB binding sites would take time and allow wasteful overexpression of many genes. Such delays, however, could be reduced by means of active molecular stripping of NF-κB from the DNA by IκB. Molecular stripping, the article points out, could solve the timescale problem that bedevils traditional thermodynamics-only models.
“The function of the NF-κB/IκB/DNA genetic switch is realized via an allosteric mechanism,” wrote the authors of the PNAS article. “Molecular stripping occurs through the activation of a domain twist mode by the binding of IκB that occurs through conformational selection.”
In addition, the authors assert that their free energy calculations for DNA binding show that the binding of IκB not only results in a significant decrease of the affinity of the transcription factor for the DNA, but also kinetically speeds DNA release.
NFκB proteins are a family of dimeric (two-part) proteins that bridge extracellular signals and gene expression, alighting on DNA to activate the manufacture of specific proteins in response to outside stimuli. They organize many cellular functions, including the inflammatory response, immune response to infection and the programmed cell death that inhibits cancer.
Because NFκB proteins can activate so many processes at once, timing is important. “NFκB isn't just sending out one signal, turning on the manufacture of one new protein. It's running a broadcast network, turning on many genes, including IκB,” said Dr. Wolynes. “The stripping mechanism means that instead of leaving all the genes turned on and letting each one figure out when to turn off, IκB strips the NFκB and makes sure they all get turned off. Transcription ends at that point.”
Dr. Wolynes and colleagues used an algorithm called AWSEM algorithm (for associative memory, water-mediated, structure and energy model) to analyze how molecular stripping works for the NFκB family. The algorithm is able to predict how proteins fold based on the interacting energies of their components.
The investigators’ molecular dynamics simulations showed that an NFκB protein attached to DNA becomes twisted when it is joined by an IκB molecule to form what they call a transient ternary structure. The structure is short-lived because the twist quickly releases the DNA.
“In the classical picture of gene switches, we don't talk about time,” Potoyan said. “We just understand that a gene turns on and off. But here, timing is critical because there are hundreds of genes being regulated. In molecular stripping, all the transcription sites become visible to IκB, which broadcasts the signal for every NFκB to unbind at once.”
Dr. Wolynes said it's known that misregulation of IκB or NFκB can lead to many medical problems, including cancer, “so turning genes off at the right time is important.” He suggested that because NFκB switches have such wide influence, targeting the switches with drugs to control specific processes will be a challenge, but one well worth pursuing.
“We believe this stripping process may be very general for master regulator genes,” he said. “It's a system that violates that 40- to 50-year-old paradigm of how a gene switch works. That's one of the reasons we think this study is important.”



