Supathep Tansirichaiya UCL Eastman Dental Institute, University College London
Liam J. Reynolds UCL Eastman Dental Institute, University College London
Gianmarco Cristarella UCL Eastman Dental Institute, University College London
Li Chin Wong UCL Eastman Dental Institute, University College London
Kimie Rosendahl UCL Eastman Dental Institute, University College London
Adam P. Roberts UCL Eastman Dental Institute, University College London
Metagenomics Helps Identify Oral Microbiome Resistance Mechanisms
Antimicrobial resistance is common in the microbial inhabitants of the human oral cavity. Antimicrobials are commonly encountered by oral microbes as they are present in our diet, both naturally and anthropogenically, and also used in oral healthcare products and amalgam fillings. We aimed to determine the presence of genes in the oral microbiome conferring reduced susceptibility to common antimicrobials. From an Escherichia coli library, 12,277 clones were screened and ten clones with reduced susceptibility to triclosan were identified. The genes responsible for this phenotype were identified as fabI, originating from a variety of different bacteria. The gene fabI encodes an enoyl-acyl carrier protein reductase (ENR), which is essential for fatty acid synthesis in bacteria. Triclosan binds to ENR, preventing fatty acid synthesis. By introducing the inserts containing fabI, ENR is likely overexpressed in E. coli, reducing the inhibitory effect of triclosan. Another clone was found to have reduced susceptibility to cetyltrimethylammonium bromide and cetylpyridinium chloride. This phenotype was conferred by a UDP-glucose 4-epimerase gene, galE, homologous to one from Veillonella parvula. The product of galE is involved in lipopolysaccharide production. Analysis of the E. coli host cell surface showed that the charge was more positive in the presence of galE, which likely reduces the binding of these positively charged antiseptics to the bacteria. This is the first time galE has been shown to confer resistance against quaternary ammonium compounds and represents a novel, epimerase-based, global cell adaptation, which confers resistance to cationic antimicrobials.
Introduction
Antimicrobial resistance is one of the world's major public health problems. Antimicrobials, including antiseptics, have been introduced and are heavily used in various industries and products such as soap, mouthwash, and detergents.1 All uses of antimicrobial agents, whether appropriate or not, increase the selective pressure for resistance. The human oral cavity is an environment that is constantly exposed to antimicrobials, for example in food and oral healthcare products, and the resident oral bacteria develop resistance to antimicrobials.
It is estimated that more than half of oral bacteria cannot be grown in a laboratory environment.2 Therefore, to analyze the entire metagenome for resistance, we can clone it into a replicon in a heterologous bacterial host such as E. coli and screen for gain of phenotype. Functional metagenomics is a powerful technique for the identification of antibiotic resistance genes, which have been performed on various environmental samples such as soil, water, and the human microflora.3–5 Novel antibiotic resistance genes have also been isolated by using functional metagenomics, for example, the chloramphenicol efflux gene pexA from Alaskan soil, a novel gentamicin resistance gene from river sediment samples, 10 novel β-lactamase gene families from the human gut microbiome, the tetracycline resistance gene tet(37), and the tetracycline and tigecycline resistance gene tetAB(60) from the human oral cavity.4–8
In this work, we aimed to detect antimicrobial resistance genes from the human oral metagenome. We chose a selection of antimicrobials, with different mechanisms of action, which oral bacteria are likely to come into contact with regularly. These include metals present in amalgam fillings, antiseptics widely used in oral healthcare products, and preservatives used in food production.
We have identified individual clones exhibiting reduced susceptibility to triclosan, cetylpyridinium chloride (CPC), and cetyltrimethylammonium bromide (CTAB) and determined the likely mechanisms responsible.
Materials and Methods
Construction of human oral metagenomics library
Human oral metagenomics DNA was extracted from the saliva samples collected from 11 healthy individuals, both males and females, from the Department of Microbial Diseases, University College London (UCL) Eastman Dental Institute. None of the volunteers had received antibiotics for at least three months before the saliva collection date. This project received ethical approval from UCL Ethics Committee (project number 5017/001), and written consent forms were collected from the volunteers. The oral metagenome was extracted as described previously.9 The library was then constructed with pCC1BAC vector and transformed into TransforMax EPI300 Electrocompetent E. coli and individual clones stored in microtiter plates at −80°C, as described previously.7
Library screening and minimum inhibitory concentration determination
The minimum inhibitory concentration (MIC) of antimicrobial compounds was determined against E. coli EPI300 containing pCC1BAC vector to be used as the concentrations for the screening. The MIC determination was performed by following the broth microdilution method described previously.10 The bacterial strains were cultured in Luria-Bertani (LB) broth containing chloramphenicol (12.5 μg/ml) and incubated overnight in 37°C shaker. The overnight culture was diluted to OD600 of 0.1. In a 96-well plate, 90 μl of LB broth, containing 12.5 μg/ml chloramphenicol and different concentration of antimicrobials (listed in Table 1), and 10 μl of diluted E. coli were added to the wells to make a total 100 μl volume. The plates were then incubated on a shaker at 37°C for 18 hr and checked for their growth to determine the MIC breakpoints.
Each 96-well plate of the human oral metagenomic DNA library E. coli was then screened against various antimicrobial substances with respect to the determined MIC. An ethanol sterilized 96-pin replicator was used to inoculate the E. coli library into a 96-well plate containing 100 μl of LB broth containing chloramphenicol and antimicrobials. The inoculated plates were then incubated in a shaking incubator at 37°C overnight.
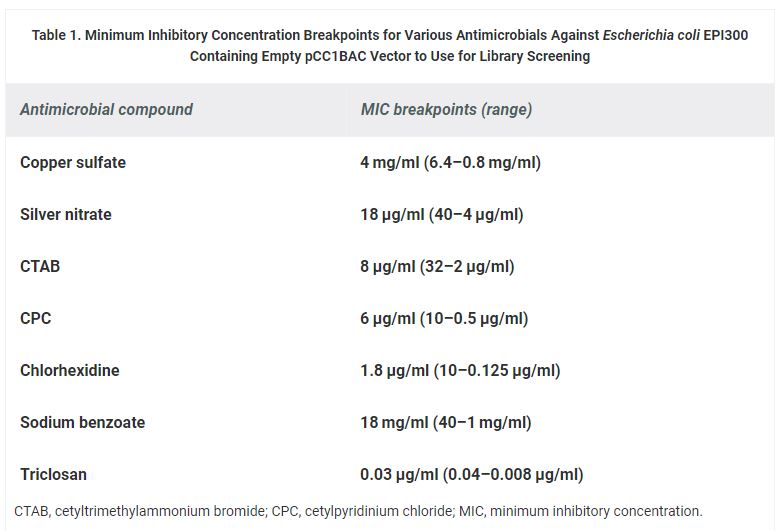
Table 1. Minimum Inhibitory Concentration Breakpoints for Various Antimicrobials Against Escherichia coli EPI300 Containing Empty pCC1BAC Vector to Use for Library Screening
Plasmid extraction and sequencing
Clones of interest were subcloned into 5 ml of LB broth containing chloramphenicol and the antimicrobials that they showed resistance to, and incubated in a 37°C shaker for 18 hr. The copy number induction culture was then set up to induce the number of the pCC1BAC plasmid in each cell from a single copy up to 25 copies per cells by transferring 1 ml of overnight culture into 9 ml of LB broth supplement with chloramphenicol (12.5 μg/ml) and CopyControl™ Induction Solution (10 μl in 10 ml), and then incubated in a 37°C shaker for 5 hr. Plasmids were extracted by the QIAprep Spin Miniprep kit (Qiagen, United Kingdom) following the manufacturer's instructions. The size of inserts was estimated by digesting the plasmids with HindIII restriction enzyme (New England Biolabs, United Kingdom), following the manufacturer's instructions, and the DNA fragments were visualized on 1% agarose gel stained with ethidium bromide. The inserts on the extracted plasmids were initially sequenced from both ends with pCC-F and pCC-R primers by using the service from Genewiz (Genewiz, United Kingdom). Additional primers were designed and used for the subsequent sequencing to extend the sequences of the inserts (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/mdr).
Transposons mutagenesis
To locate the genes conferring resistance, transposon mutagenesis was performed by using the Template Generation System II Kit (Thermo Scientific, United Kingdom), following the manufacturer's instructions. Entranceposon (KanR-3) was selected for the mutagenesis. The reaction mixture was electroporated into E. coli EPI300 (Cambio, United Kingdom) and grown on LB agar with chloramphenicol (12.5 μg/ml) and kanamycin (20 μg/ml). Each colony was then subcultured into 96-wells plates containing LB broth containing chloramphenicol (12.5 μg/ml), kanamycin (20 μg/ml), and the corresponding resistant antimicrobials (8 μg/ml CTAB for A10F2 and 0.06 μg/ml triclosan for C7C4). After incubating overnight in a 37°C shaker, clones that lost the resistant phenotype were selected for the extraction of their plasmids and sent for sequencing with SeqE and SeqW primers, provided with the kit to determine the target sites.
Detection of fabI on the DNA inserts of triclosan-resistant clones
The presence of the fabI gene on the DNA inserts of each triclosan-resistant clone was carried out by using a rational polymerase chain reaction (PCR) approach. Primers were designed to target fabI gene from different bacterial species, identified by the end sequencing of each clone (Supplementary Table 1). The PCR reaction contained 15 μl of 2 × BioMix Red (Bioline, United Kingdom), 0.2 μM of each primer, 50–100 ng of extracted plasmid from triclosan-resistant clones, and molecular grade water (Sigma, United Kingdom) up to 30 μl. The PCR cycle was programmed as follows: initial denaturation at 94°C for 3 min, 35 cycles of (i) denaturation at 94°C for 1 min, (ii) annealing at 54–58°C for 30 sec, and (iii) extension at 72°C for 1 min, and final extension at 72°C for 3 min. The PCR products were then purified using the QIAquick PCR Purification Kit (Qiagen, United Kingdom), following the manufacturer's protocols, and confirmed by sequencing.
Subcloning of UDP-glucose 4-epimerase and glucose-6-phosphate isomerase genes
The primers, listed in Supplementary Table 1, were designed to amplify the UDP-glucose 4-epimerase and glucose-6-phosphate isomerase (GPI) genes, identified from the transposon mutagenesis. The PCR reaction contained 15 μl of 2 × BioMix Red (Bioline, United Kingdom), 0.2 μM of each primer, 50–100 ng of A10F2 plasmid, and molecular grade water (Sigma, United Kingdom) up to 30 μl. The PCR cycle was programmed as follows: initial denaturation at 94°C for 3 min, 35 cycles of (i) denaturation at 94°C for 1 min, (ii) annealing at 54–58°C for 30 sec, and (iii) extension at 72°C for 1 min, and final extension at 72°C for 3 min. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, United Kingdom), following the manufacturer's protocols. The amplicons were digested with EcoRI and HindIII restriction enzymes (New England Biolabs, United Kingdom) and directionally cloned into predigested pCC1BAC vector. The ligation mixture was electroporated into electrocompetent E. coli EPI300 and grew on LB agar supplemented with chloramphenicol (12.5 μg/ml), 0.4 mM IPTG, and 40 μg/ml Xgal. Clones with inserts were selected and confirmed by extracting the plasmids and sequencing across the cloning site.
To read this article in its entirety and access its referencs click here.
Microbial Drug Resistance, published by Mary Ann Liebert, Inc., is an international, peer-reviewed journal that covers the global spread and threat of multi-drug resistant clones of major pathogens that are widely documented in hospitals and the scientific community. The above article was first published in the March 1, 2018 issue of Microbial Drug Resistance with the title “Reduced Susceptibility to Antiseptics Is Conferred by Heterologous Housekeeping Genes". The views expressed here are those of the authors and are not necessarily those of Microbial Drug Resistance, Mary Ann Liebert, Inc., publishers, or their affiliates. No endorsement of any entity or technology is implied.







