Anton Simeonov Ph.D. National Institute of Health
This Article Discusses a Robust Regulator of G Protein Signaling (RGS) Inhibitor Discovery and Characterization Paradigm
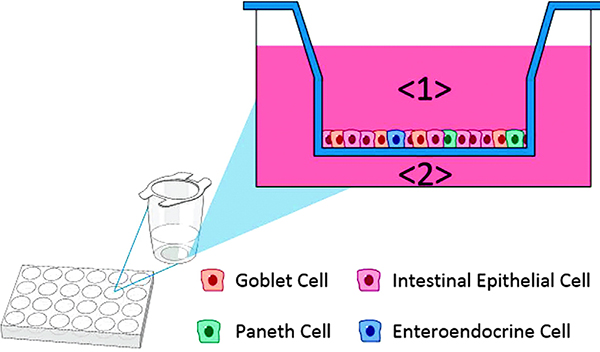
The Microphysiological Systems program (also known as tissues-on-chip, organs-on-chips, or tissue-chip) represents a major effort by several U.S. government agency partners to advance the development and validation of platforms recapitulating human physiology in microfluidic systems for the purpose of drug discovery and toxicity testing in an experimental environment, which provides an improved predictive power relative to the currently utilized cellular and animal models. To accomplish this task, it is essential to produce the various components of the human physiology, that is, the organs, and demonstrate their functionality, and then effectively connect the various organs into a human organism–like system in order to recapitulate the more complex physiological processes. To date, nearly a dozen human organ systems have been produced and validated by a consortium of almost two dozen universities, nonprofit institutes, and private partners, and several organ systems have been linked together to begin to model complex physiological processes. At present, the program is directing resources toward external testing and validation of the organs-on-chip by placing the systems into labs different from those that created them in order to demonstrate real-life adoption. The present paper is a wonderful demonstration of the level of complexity of human physiology that can be interrogated through these systems. Here, a multi-institution team connects four models (Figure 1): the human intestine (Johns Hopkins University [JHU]/Baylor College of Medicine [Baylor]), the sequentially layered self-assembly liver (SQL-SAL, University of Pittsburgh [UPitt]), a vascularized or non-vascularized proximal tubule kidney (VPTK or PTK, University of Washington [UWash]), and an intact blood–brain barrier/neurovascular unit (BBB/NVU, Vanderbilt University). These organ systems were selected for their utmost importance during the preclinical development of drug candidates. The team achieved the functional coupling of the four organ systems, while also allowing for the collection of a wealth of validation data (Figure 2). Uptake, transport, metabolism, and side effects of several drug molecules were successfully demonstrated in the integrated system, marking the beginning of a new and very exciting phase in the development of a “human-on-a-chip.”

Figure 1. (B) Media from the jejunum intestine basolateral compartment <2> is perfused as a 1:3 jejunum/naïve liver media into the influx port of the SQL-SAL liver model <3>. Efflux media is collected <4> and used to add to two downstream organ models.
Figue 1. (C) The vascularized kidney proximal tubule module is a two lumen, dual perfusion system. For the vascular compartment, jejunum/liver-conditioned media <4> is diluted 1:2 or 1:4 with naïve EGM-2 media and then perfused into the influx port <5> to collect effluent from the proximal tubule at <6>. In parallel with perfusion through the vascular compartment, the proximal tubule compartment is perfused with naïve DMEM/F12 PTEC media <6> for effluent collection. (D) The blood–brain barrier with NVU is constructed in a membrane-separated, two-chambered microfluidic device. The brain-derived endothelial vascular compartment is perfused at the influx port <7> with jejunum/liver-conditioned media <4> diluted 1:4 with naïve EGM-2 media. The effluent is collected at the efflux port <7>. In parallel with perfusion through the vascular compartment, the neuronal cell compartment is perfused with naïve EBM-2 media at the influx port <8> for effluent collection at <8>.
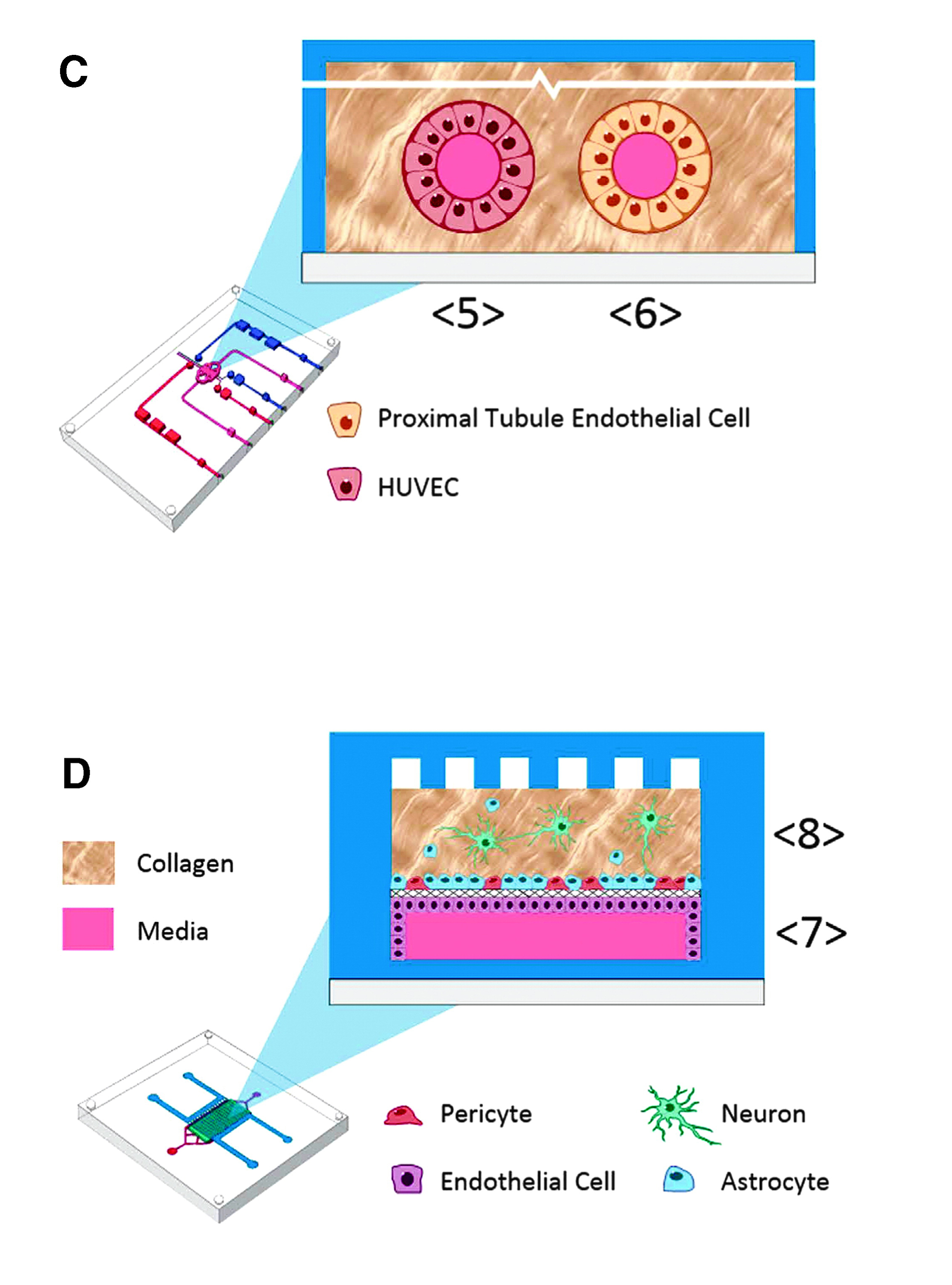
Figure 2. Work flow for functional coupling experiments. Terfenadine exposure is used as an example; TMA and vitamin D3 experiments followed essentially the same workflow. The test compound is initially added to the apical gut media and samples are collected from the apical and basolateral media for MS analysis. Basolateral media samples are sent to UPitt where they are mixed with liver media for exposure in the liver module. Effluent samples are taken for MS analysis and sent to UWash and Vanderbilt where they are mixed with kidney and NVU media, respectively. Samples are taken of the effluent from the kidney proximal tubule module and from the vascular and brain sides of the NVU for MS analysis.

* Abstract from Sci Rep 2017;7:42296
Organ interactions resulting from drug, metabolite or xenobiotic transport between organs are key components of human metabolism that impact therapeutic action and toxic side effects. Preclinical animal testing often fails to predict adverse outcomes arising from sequential, multi-organ metabolism of drugs and xenobiotics. Human microphysiological systems (MPS) can model these interactions and are predicted to dramatically improve the efficiency of the drug development process. In this study, five human MPS models were evaluated for functional coupling, defined as the determination of organ interactions via an in vivo-like sequential, organ-to-organ transfer of media. MPS models representing the major absorption, metabolism and clearance organs (the jejunum, liver and kidney) were evaluated, along with skeletal muscle and neurovascular models. Three compounds were evaluated for organ-specific processing: terfenadine for pharmacokinetics (PK) and toxicity; trimethylamine (TMA) as a potentially toxic microbiome metabolite; and vitamin D3. We show that the organ-specific processing of these compounds was consistent with clinical data, and discovered that trimethylamine-N-oxide (TMAO) crosses the blood-brain barrier. These studies demonstrate the potential of human MPS for multi-organ toxicity and absorption, distribution, metabolism and excretion (ADME), provide guidance for physically coupling MPS, and offer an approach to coupling MPS with distinct media and perfusion requirements.
Anton Simeonov, Ph.D., works at the NIH.
ASSAY & Drug Development Technologies, published by Mary Ann Liebert, Inc., offers a unique combination of original research and reports on the techniques and tools being used in cutting-edge drug development. The journal includes a "Literature Search and Review" column that identifies published papers of note and discusses their importance. GEN presents here one article that was analyzed in the "Literature Search and Review" column, published in Scientific Reports titled "Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood–brain barrier and skeletal muscle", authors is Lawrence Vernetti.