July 1, 2014 (Vol. 34, No. 13)
Shawn C. Baker Ph.D. co-founder and CSO AllSeq
Next-generation sequencing (NGS) has been making tremendous strides in the research market and with Illumina’s recent launch of the HiSeq X Ten, we’ve essentially reached the $1,000 genome (notwithstanding quibbles over what exactly should be accounted for in the $1,000). With these advancements, the pull to adapt NGS for the clinical market has gotten stronger. The first examples of how this technology will transform medicine are already showing up.
The press has been filled with stories, sometimes heart-warming and sometimes gut wrenching, of NGS being used in the diagnosis of late-stage cancers and rare childhood diseases. But while these examples show how NGS can be used to treat patients, they aren’t really examples of clinical sequencing. These are still research projects, each one requiring the attention of multiple experts poring over the data to come up with hypotheses of how to treat the underlying malady. In order for NGS to be considered a true clinical tool, it will have to be used on a routine basis, replacing older genetic-based tests. This transition is starting to happen for prenatal testing and cancer diagnostics.
One of the critical requirements for next gen sequencers for the clinical market is stability. The platforms must operate reliably day in and day out without any changes or modifications. [Maggie Bartlett, NHGRI]
The High-Throughput Trend
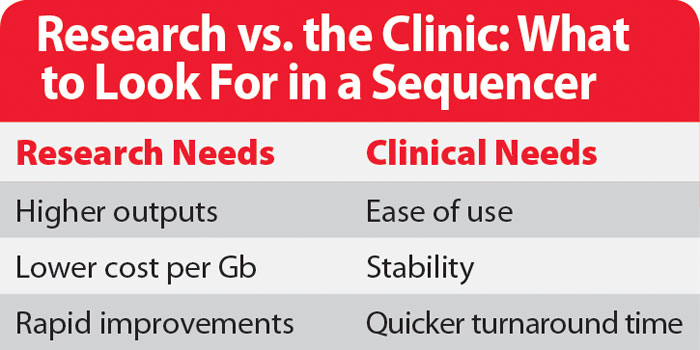
In the infancy of the NGS market, early adopters really only cared about two things: how much raw output the new machines could generate and how quickly the price per base could drop. Secondary to these two drivers was the pressure to increase the read length back up to the standard of Sanger sequencing, the mainstay that NGS was displacing. These driving goals, combined with the intense competitive pressure seen in the market between 454, Illumina, Life Technologies, and Complete Genomics, led to incredible output gains and an equally incredible drop in price, with Illumina’s flagship HiSeq X generating nearly 2 Tb of data over 3 days at a cost of only $7 per Gb.
The Desktop Trend
While the cost of generating a Gb of data had been steadily dropping, the cost of each individual run had actually been climbing. A full run on Illumina’s HiSeq 2000 costs over $20,000. And the capital expenses are quite high—up to $750k. With the announcement of Ion Torrent’s PGM, however, the market started moving in a different direction. The PGM launched with a sticker price of just $50k (rounding up to $75k or so with ancillary equipment) and a per-run cost of just $1,000. Illumina quickly followed suit with the launch of the MiSeq—$125k machine and sub-$1,000 runs.
The Clinical Trend
In addition to cheaper instruments and cheaper runs, the desktop instruments introduced two other features that are critical for the clinical market. The first is speed, which is where the PGM really shines. With short runs of less than four hours, the PGM makes it possible to reduce the turnaround time to less than a day—a critical feature when dealing with patient samples. The MiSeq started the other critical trend—a substantially simplified workflow. While the library prep is still performed “off instrument,” the rest of the steps are handled in an automated fashion with the push of a few buttons. This even extends to data analysis as each MiSeq is connected to BaseSpace, Illumina’s online data analysis solution.
The other major need of the clinical market is stability—the platforms need to operate reliably day in, day out without any changes or modifications. And this is perhaps the biggest departure from the research market. Changes and improvements to NGS platforms happen all the time. Customers have been tolerant of changes, even expecting or demanding them all in the quest for increased performance specs. But clinical customers are willing to give up steady improvements for the sake of stability. The regulatory hurdles involved in getting a platform or diagnostic cleared for clinical use are arduous, and every change to the system requires a repeat of at least part of that process. While systems that have been cleared by the FDA can change and improve over time, the pace is substantially slower.
MiSeqDx—First Clinical NGS System
In November 2013, Illumina achieved a historic milestone in next-generation sequencing when its MiSeqDx sequencer was approved by the FDA for use in clinical settings. Specifically, this instrument, a modification of their standard MiSeq, achieved 510(k) clearance in conjunction with two assays for cystic fibrosis along with a universal library kit that will enable users to create their own assays for this platform.
Although the MiSeqDx is based on the original MiSeq platform, the two systems have now started to diverge. While the original research platform has seen its chemistry upgraded to the latest v3 kits (capable of generating up to 15 Gb of output per run), the MiSeqDx was cleared using the original v1 chemistry, so the output is currently only around 1 Gb per run. While this may climb over time, technology advancements on this platform will be substantially slower—stability has a price.
Future Platforms
In launching this system, Illumina has paved the way for future FDA-cleared systems, both from Illumina and its competitors. As the first NGS system cleared by the FDA, the MiSeqDx has become the “platform of reference” against which future platforms will be compared. These future platforms will still have to go through clinical trials, but the hurdle will be a bit easier as they just have to show “substantial equivalence” to the MiSeqDx.
Based on the success they’ve had so far, Illumina has announced that they’ll also submit their high-throughput platform, the HiSeq 2500, for FDA clearance. Following a similar strategy, they will release it with a specific assay (in this case it is likely to be based on their verifi® prenatal test) along with a universal kit.
Not to be outdone, Life Technologies (now part of Thermo Fisher Scientific) has announced that it will be submitting its Ion Torrent PGM platform for FDA clearance in the fall of 2014. This platform has already proved popular with clinical researchers due to its speed; a rapid turnaround time can be critical when dealing with patient samples. One area that the Ion Torrent platforms have lagged is that of ease of use due to their reliance on emulsion PCR, a tricky and time-consuming process. In an effort to address this issue, Life Technologies has launched the IonChef, an automated system that handles both emulsion PCR and chip loading.
Because NGS for the research market is dominated by Illumina, and also by Thermo Fisher Scientific to a smaller extent, developers of new platforms are focusing primarily on clinical sequencing, hoping to get a toe-hold in what is projected to be a large and fast-growing market.
Qiagen has plans to launch its new system, the GeneReader, later this year. While it uses a sequencing chemistry similar to that of Illumina, it will cater to the clinical user by using a larger number of small output flow cells, reducing or even eliminating the need to pool samples, which could lead to a quicker turnaround time and less cross-contamination risk. Also, the GeneReader will be integrated into a comprehensive workflow from Qiagen, covering sample prep all the way through data analysis.
Bio-Rad Laboratories recently entered the NGS market through its acquisition of GnuBIO. The GnuBIO targeted sequencing platform, which is currently in beta testing, uses picoinjector technology to form droplets in which individual sequencing-by-hybridization reactions take place. In addition to reducing reagent volumes (which keeps the price down), the microfluidic droplet system allows the user to generate arbitrary amounts of data that are tailored to their particular needs rather than generating a set amount of data for each run. This flexibility should fit well with the clinical workflow.
The Road Ahead
Clinical next-generation sequencing is making rapid strides, but there is still a long way to go before it becomes routinely used with patients. Currently there is only a single FDA-cleared clinical NGS platform on the market, and it will take at least a couple of years before it faces serious competition. Also, turning the raw output of the sequencers into a report that clinicians and physicians can use to make decisions on how to treat their patients remains a challenge. Finally, NGS is progressing technologically at a pace that the regulatory and ethical bodies are finding difficult to match. So while the $1,000 genome is finally here, widespread adoption in the clinic is still in the future.
Shawn C. Baker, Ph.D. ([email protected]), is the CSO and co-founder of AllSeq.