November 15, 2014 (Vol. 34, No. 20)
MaryAnn Labant
Toxicity testing may feel like walking a tightrope, but a slip needn’t kill development, not with better assays, chemoinformatics, and risk/benefit analyses.
The issue of drug safety is as old as drug development itself. Historically, animal models have been the cornerstone of preclinical drug development, resulting in mostly phenomenological observations rather than providing a deeper mechanistic understanding of the side-effects of drugs or compounds in development.
As modern techniques progress, more predictive assays may allow safety pharmacologists to perform better informed risk assessment.
A major reason for drug liability and withdrawal has been cardiovascular side-effects, primarily drug-induced Torsades de Pointes (TdP), a potentially fatal form of ventricular arrhythmia. The major preclinical assay required to gauge the risk of compounds for TdP is the hERG assay, which is not sufficiently predictive because it tends to produce many false-positive results.
According to Yama Abassi, Ph.D., vice president of ACEA Biosciences, the hERG assay’s poor predictive value means that a number of good compounds in clinical development are being discontinued. The industry urgently needs sufficiently predictive assays that can screen compounds in fairly high throughput and also provide mechanistic toxicity information.
The availability of human induced pluripotent stem cell (iPSC)-derived cardiomyocytes in scaleable quantities for screening purposes has generated excitement as a relevant and predictive model system. The challenge is deriving pertinent information from these cells for risk-assessment purposes.
ACEA’s new xCELLigence® CardioECR system combines two cutting-edge technologies, impedance and field potential electrodes, in the same platform. The impedance electrodes allow for assessment of cardiomyocyte contractility and viability, while the field potential electrodes measure integrated ion channel activity.
The system conducts both readouts simultaneously to capture and measure the events known as excitation-contraction coupling. The parameters and algorithms in the accompanying software identify and analyze interesting aspects of cardiomyocyte response to compound treatment.
Furthermore, the FDA has begun to take notice of the importance of in vitro preclinical assays for screening of proarrhythmic compounds. The agency is working to establish the Comprehensive in vitro Proarrhythmia Assay (CiPA), a novel safety screening paradigm. Through the utilization of high-throughput, predictive, and mechanistic assays, CiPA attempts to identify proarrhythmic compounds earlier in the drug discovery process.
Currently, the CiPA stakeholders are working together to design well-controlled validation studies to assess the reproducibility of the assays across different instruments and different cells. A multisite study may be completed as early as this month.

The xCELLigence RTCA CardioECR system from ACEA Biosciences combines impedance and multi-electrode array (MEA) technology with a pacing function. The system is designed to allow simultaneous real-time measurement of cardiomyocyte contractility by impedance, and electric activity by field potential measurement.
Liver Toxicity
A CiPA-like path is not yet in place to address the risk of rare or idiosyncratic drug-induced liver injury (DILI). Increasingly, expanded clinical studies are needed to rule out such risks. In general, today’s drugs will not cause a liver problem in animal studies, yet there are still liver problems in humans.
“People say we have to humanize preclinical testing. But even humans are not a good model for this rare toxicity. The average human can take the drug entirely safely,” discussed Paul Watkins, M.D., director, Hamner-University of North Carolina Institute for Drug Safety Sciences.
“The chance that you are going to find liver cells to put into a mouse or to culture that are indicative of a rare event is low,” Dr. Watkins continued. “The problem is that liver cells available now are from random donors. If you could pick someone that actually had this rare event, and use iPSCs to make liver cells from that individual, you could at least begin to understand for that particular drug what is different in their liver cells from the average person.”
The University of North Carolina is the center for the Collaborative Cross, a population of inbred mice that have been mated according to a computer algorithm to arrive at the maximum genetic diversity. Forty-five strains are currently ready, and the complete DNA sequence of every mouse is known. The mice can be used to screen drugs, and to begin to understand the genetic determinants.
Drugs that unexpectedly failed due to DILI have been shown to present toxicity in some strains of these heterogenetic mice providing models for mechanistic studies.
The Hamner Institutes is also the seat for a major public/private partnership, the DILI-sim Initiative, which is developing a computer based-model of mechanisms that underlie liver toxicity of drugs. DILIsym® software can create virtual patient populations to predict rare liver-toxicity events, identify factors that place patient subpopulations at increased risk, and contribute to risk-management strategies.
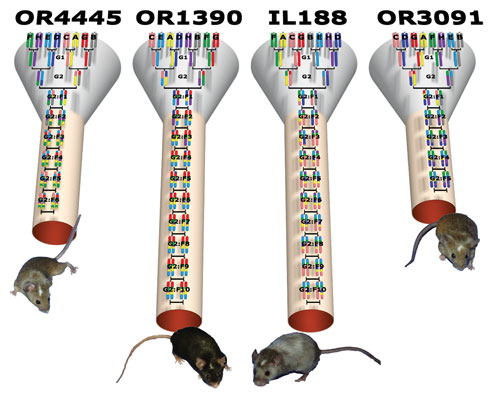
The Collaborative Cross addresses many shortcomings in available mouse strain resources, including small numbers of strains, limited genetic diversity, and a nonideal population structure. The breeding approach is designed to randomize the genetic makeup of each inbred line. [Hamner Institutes / University of North Carolina]
In Silico Predictions
Inspired by the ligand-based organization of targets in classical pharmacology, the Similarity Ensemble Approach (SEA) is a rapid, broad-scale, and model-free chemoinformatic computational method that operates on hundreds of thousands of ligands organized by their molecular targets.
“SEA allows us to ask and quantify whether any given drug or drug-like molecule is more similar to a target’s ligands than would be expected by random chance alone. A strong significance score is a prediction of possible off-target activity for that drug,” explained Michael Keiser, Ph.D., founder and chairman of SeaChange Pharmaceuticals and assistant professor at the University of California San Francisco School of Medicine.
“In collaboration with Novartis, we were able to determine that some off-targets were antitargets, which we found to be associated with adverse drug reactions (ADRs) via a guilt-by-association enrichment factor metric,” added Dr. Keiser. “Correspondingly, we were able to show that these antitargets were often better explanations of the drug’s known adverse reactions than any of its previously reported targets.”
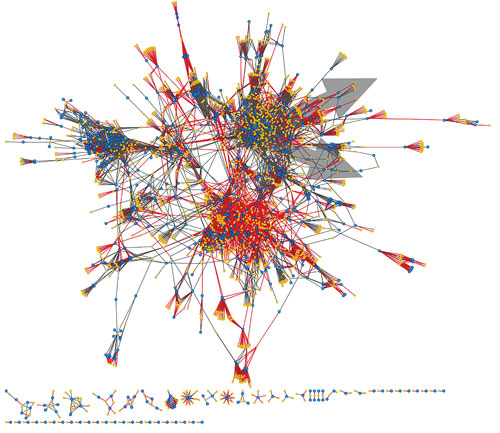
A global network of off-targets for 1,200 FDA-approved drugs. Gold nodes are drugs; blue nodes are targets; gray edges are known links; and red edges are strong, new Similarity Ensemble Approach (SEA) predictions. According to SeaChange Pharmaceuticals, SEA—a chemoinformatic computational method—can predict drug side-effects and identify new therapeutic opportunities.
Off- and antitarget predictions are intended to prioritize, not to replace, the use of screening and experimental safety assays. Calculations are run rapidly on tens or hundreds of thousands of drug-like molecules to determine areas of potential liability for compounds.
Typical positive predictive values (PPVs) are 48–52%, even among known drugs, where one might have expected previously unreported antitargets to be sparse. These PPVs are approximately an order of magnitude better than those achieved in most virtual-screening campaigns.
The approach can be used for drugs and small molecules in development or on the market with known structures, and focuses on predicting ADRs that are mediated by specific antitargets.
The future goal is direct prediction of entire multitarget combinations. The increasing availability of big data may one day allow linking a drug’s full on- and off-target profile with a patient’s unique genomic profile and electronic medical records for use in the emerging field of systems pharmacology.
Early Disease Detection
Many existing biomarkers are over 100 years old and lack the accuracy needed for timely detection of disease.
Mechanistic biomarkers of drug safety allow informed decisions about a drug’s safety profile early in the development process. In collaboration with the Predictive Safety Testing Consortium, the Vaidya Laboratory was able to take the biomarker, kidney injury molecule-1 (KIM-1), forward from evaluation to confirmation, regulatory qualification, and point-of-care diagnostic testing for commercial utility.
Although seven biomarkers, including KIM-1, were qualified for preclinical toxicity in 2008, the clinical qualification of these biomarkers remains an uphill task. Early results indicate that the list of kidney-disease biomarkers may need expansion given the heterogeneity and complexity of the patient population. MicroRNAs (miRNAs) have shown promise as potentially noninvasive and more stable biomarkers easily quantifiable in biospecimens.
“We have generated miRNA profiles in human urine from patients with and without kidney injury to identify three to five miRNAs to be early diagnostic, sensitive and specific indicators of different forms of kidney disease,” stated Vishal Vaidya, Ph.D., head of systems toxicology in the Harvard Program in Therapeutic Sciences at Harvard Medical School and assistant professor of medicine and environmental health at Brigham and Women’s Hospital and Harvard School of Public Health.
These miRNAs provide mechanistic information about the disease pathophysiology in the kidney. For example, miR-21, found to be significantly increased in patients with kidney injury, regulates metabolic alterations resulting in kidney damage. Because miR-21 is in the causal pathway of disease initiation and progression, measurement of miR-21 provides more mechanistic information.
The key is to now compare these emerging miRNA biomarkers in the urine with the candidate protein biomarkers in a large number of patients, with and without kidney disease, over time to understand the temporal profile and determine their effectiveness as early kidney-damage indicators as well as for their utility in patient stratification.
Quantitative Risk-Benefit Assessment
Quantitative risk-benefit assessment (RBA) requires a challenging combination of forward thinking and consistency of judgment. Standardized policies are required as well as industry agreement on the methodology for translation across organizations.
According to Israel Gutierrez, M.D., vice president of drug safety at Exelixis, professionals familiar with a qualitative model may harbor doubts about the validity and potential of the quantitative approach, impeding consensus. Exelixis performs quantitative RBA in an exploratory manner, on an ongoing basis, following methodology accepted by the European Medicines Agency (EMA) and the work processes put forward since 2010. The FDA is in the process of determining its policies.
There are roughly 18 quantitative approaches to RBA. Out of all of the different methods, the medical application of multicriteria decision and analysis (MCDA) is generating interest. Around since the 1970s, MCDA has been applied to drugs that have been approved, but historically it has not been easy to adopt or utilize.
New visual tools allow implementation of sensible and appealing methodologies, enabling communication of findings and analy ses to wider audiences without the need to delve into the statistical intricacies.
The assessment of benefit versus risk can change over time, as the understanding of drugs and population-specific characteristics evolves. It is now possible to project the contribution of the risk and benefit of those relationships, and the impact of risk management on high-risk events, in circumstances that are considered important by companies and regulatory agencies.
The EMA and others have already begun to assess the benefit/risk profile of drugs in their portfolio and in development. Regardless of the method used, how one applies and weighs benefit/risk requires a methodology and a plan that is applied consistently a priori. Varying methodologies can curtail and prevent progress of the analysis overall.
Biocontainment Lab to Use Provantis Toxicology Software
The University of Louisville (UofL) Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases bought Instem’s Provantis® preclinical software solution as part of an expansion program. Provantis, a fully integrated Windows-based system with applications in nonclinical toxicology evaluation studies, will be used within the Regional Biocontainment Laboratory. This state-of-the-art facility provides scientific expertise and biosafety level-three laboratories for the discovery of therapeutics for serious pathogens and an infrastructure to meet pandemic or bioterrorism emergency needs.
UofL funding for the expansion of its facilities and the purchase of Provantis were awarded through a National Institutes of Health initiative to improve the nation’s top research facilities.



