August 1, 2006 (Vol. 26, No. 14)
Addressing Cell and Protein Function Using Standard Recombinant Approaches
Until recently, the principal way to study changes in cellular protein expression levels and perform protein functional analysis was by fluorescent reporter proteins combined with automated confocal imaging systems, both of which provide the basis for high-content screening (HCS) protocols. However, the large size of fluorescent proteins (~26 kD) and relatively high background fluorescence in the cell mandate that the fusion protein be overexpressed for detection, which frequently perturbs the biology being studied.
Immunostaining fixed cells has also been widely used to visualize a protein’s sub-cellular location. However, the time-consuming, laborious labeling procedures are not as amenable to automated fluid handling. Furthermore, high-affinity, selective antibodies suitable for imaging are not always available when a new protein target is identified. These issues are compounded by the fact that HCS protocols generate large amounts of complex data requiring extensive analysis. Although HCS assays have the advantage of multiplexing to simultaneously analyze different cellular events, the infrastructure needed and high instrumentation cost are barriers to routine adoption of HCS in primary screening.
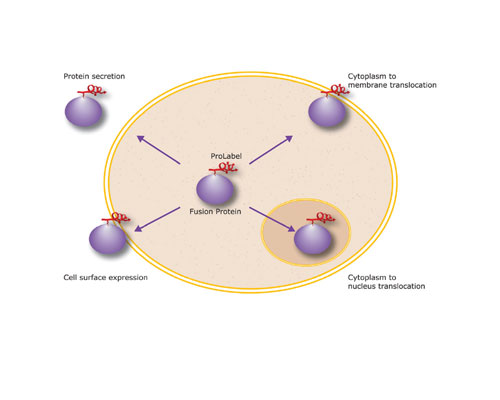
Cell-based high-throughput screening (HTS) assays thus require a technology platform for studying a variety of protein functions in a high-throughput, automated, and flexible manner. DiscoveRx (www.discoverx.com) provides a technology platform of PathHunter assays using a small ProLabel peptide tag. This tag allows the study of many cell functions in live cells, including protein expression, degradation, secretion, translocation, and trafficking, as well as phenotypic events, such as mitosis (Figure 1). Moreover, the flexibility of the ProLabel tag allows screening using one-step homogeneous assay protocols for small molecules or dissecting protein pathways by introducing siRNA into cells. The properties of the ProLabel tag thus afford a flexibility that distinguishes the PathHunter technology as a unique platform with applications in all phases of drug discovery from target identification through lead optimization (Table).
The properties of the ProLabel tag allow high-throughput interrogation of many protein functions in vivo, including quantitating the effects of siRNA in target validation, hit identification, or compound library screening. Biochemically, the ProLabel tag is a small peptide fragment of the well-characterized b-galactosidase (b-gal) enzyme. In the absence of the ProLabel tag, the large b-gal fragment called Enzyme Acceptor (EA) is inactive. However, when the ProLabel peptide combines (complements) with EA, active β-gal is formed.

Fig 1: Sub-cellular localization of ProLabel-tagged proteins provides a versatile functional activity marker
The ProLabel Peptide Tag
The ProLabel peptide can be fused to virtually any protein, yet retain the ability to complement with EA. Adding a solution of chemiluminescent substrate to the assay generates a signal detectable in microtiter plates using a simple luminometer. The process of combining the ProLabel tag with EA is termed Enzyme Fragment Complementation (EFC)a technique used in numerous biochemical screening assays developed and commercialized by DiscoveRx. The flexibility of EFC is further demonstrated in several applications designed to detect protein movement within the cell. To accomplish this, ProLabel and EA fusion protein constructs are made with specific localization or signaling sequences to direct their expression to specific sub-cellular compartments. When a cellular pathway is activated, the fusion protein translocates, delivering the ProLabel tag from the cytoplasm to an intra- or extra-cellular destination, such as the nucleus, inner plasma membrane, or cell surface, where EA is targeted. This results in compartment-specific signal generated by EFC, i.e., positional complementation.
Cell-based Functional Assays
Several PathHunter assays for cell signaling, protein degradation, translocation, and proliferation have been introduced by DiscoveRx for use in many areas of drug discovery. These include target identification, target validation, primary screening, lead compound optimization, and toxicology.
Since the assays are homogeneous, antibody-free, and designed for primary screening in 96-, 384-, or 1536-well microtiter plates, they are suited for high-throughput automation. All of them also have high sensitivity provided by catalytic generation of a chemiluminescent signal arising from high levels of b-gal substrate hydrolysis.
Depending upon the biological pathway of interest, different PathHunter assay formats can be used. Some have cells expressing EA in a discrete cellular compartment, like the nucleus, to assess translocation of proteins. For example, nuclear translocation of a ProLabel-glucocorticoid receptor fusion can be measured in a homogeneous fashion without imaging. Translocation is detected when the ProLabel fusion protein moves into the nucleus upon activation, generating signal.
Similar assays have been developed using a range of nuclear hormone receptors and other key signaling molecules, such as NFAT. Other assay formats involve adding EA as an exogenous reagent to a cell lysate or whole cells containing ProLabel fusion proteins. Protein expression, secretion, or degradation can be measured in this way.
Measuring RNA Interference
Discovering highly selective siRNA oligomers that knockdown the expression of a protein from a target gene involved in cellular signaling is a widely used approach in drug discovery. However, identifying active siRNAs requires screening of siRNA libraries using cell-based assays in which changes in protein function can be determined rapidly and quantitatively.
This need for a convenient screening technology to assess in vivo siRNA effects led to use of the PathHunter technology platform for siRNA screens. For example, the PathHunter Mitotic Index Assay can be used to screen siRNA to identify novel anticancer targets. In this microtiter-based assay, EA is stably expressed in the nucleus and a ProLabel-tagged protein is expressed only in the cytoplasm. When the nuclear envelope is degraded during the mitotic phase of cell division, EFC occurs, resulting in chemiluminescent signal that increases proportionately to the mitotic activity.
Here we illustrate the potential for screening of siRNAs to assess the function of genes regulating proliferation (Figure 2). PathHunter HEK 293 Mitotic Index Cells were transfected with KIF11, NUMA1, and CDK6 siRNAs. These oligomers target genes known to block mitosis. Following validation of gene knockdown by Western blotting, PathHunter Assays were run. The results were consistent with decreased protein expression. However, the homogeneous HTS format made the assay faster and simpler than traditional HCS or Western blotting.

2a: Western blot of target protein expression and corresponding siRNA knockdown
Versatile Assays
In summary, the PathHunter assays, employing the ProLabel tag, impart versatility to screening and assay development groups preferring a single assay platform. Investigators can now address many aspects of cell and protein function using standard recombinant approaches, without expensive confocal imaging instruments and complex data analysis. Since the small, inert ProLabel tag does not interfere with protein function, one canwithout over-expression of the target proteingenerate a highly amplified chemiluminescent signal, enabling simple, cost-effective detection in a biologically relevant environment. DiscoveRx offers many ready-to-go PathHunter Biosensor assays for primary screening. To aid investigators in assay development, many PathHunter Cell Lines and ProLabel vectors are available.

2b: PathHunter EFC Assays demonstrate correspondng knockdown patterns