October 15, 2009 (Vol. 29, No. 18)
Sophia Fox
Academia Paves the Way with More Portable, Reproducible, and User-Friendly Tools
A team from the Technical University of Denmark and the Copenhagen University Hospital recently published research results obtained with a microfluidic device known as a Multi-Stringency Array Washer (MSAW). According to the researchers, the MSAW facilitates probe optimization and, therefore, leads to increases in both microarray sensitivity and specificity. Using the MSAW also negates the need for extensive assay optimization and time-consuming thermal gradient/melting curve approaches, as reported at Select Bioscience’s “Microarray World Congress” held recently in San Francisco.
“Because microarrays are vast multiplex assays, potentially containing thousands of different probes, assay design relies on researchers’ ability to predict properties such as the melting temperature of every probe on the array,” explained Martin Dufva, Ph.D., associate professor at the Technical University of Denmark.
“Thermal gradients and their resulting melting curve data can be used to help produce meaningful and clear results from microarrays. The use of temporal or spatial temperature gradients to create melting curves has a number of drawbacks, however, including complexity of array fabrication, array size limitations, sample throughput, heat dissipation, and incompatibility with scanner instrumentation.”
The Danish researchers suggested that differential stringencies created by buffers of varying ionic strength represent a more useful, easily implementable, and flexible alternative to thermal gradients for assay development. The MSAW, which was developed to apply this concept, flows buffers of different ionic strength over different regions of a single microarray.
“We have published studies in which the MSAW was used with a microarray designed to genotype small variations in the human beta-globin gene, under different ionic strengths,” Dr. Dufva continued. “The results confirmed that an ionic gradient functions much like a temperature gradient, resulting in dissociation curves similar to those produced by discrete temperatures.
“Importantly, and in contrast to thermal-gradient technologies, the MSAW only requires washing for a fixed period, scanning is carried out at one temperature using dried slides, and there is no need for multiple exposures that can cause photobleaching. And, in contrast to multithermal technologies, the MSAW is adaptable to commercial high-density arrays as well as custom-made slide layouts.”
The MSAW developed by Dr. Dufva’s team is, in fact, a reincarnation of a multi-thermal array washer they previously developed. Whereas the multithermal array washer created eight individually controlled heating zones, each corresponding to a subarray, the MSAW creates eight zones that allow post-hybridization washing of each of the subarrays at different ionic strengths.
The MSAW device consists of a solid support onto which an elastomeric layer containing microfluidic chambers and channels is mounted. The chambers separate the slide into distinct regions, each of which can be washed under a different stringency condition. The glass slide containing the microarray is positioned via alignment features within the device and the assembly is sealed using a lid that applies pressure.
“It’s a convenient tool for assay development because we can test probe function empirically on a single slide, rather than having to run multiple slides at different stringency conditions or temperatures,” Dr. Dufva pointed out. “In addition to its use in probe design, the approach can also form part of the assay itself, allowing researchers to use combinations of probes on one slide that would generally not be feasible if assayed under a single stringency.”
The team has used the MSAW in a multiparametric study evaluating the hybridization of DNA to probes with different GC content and with different probe and spacer lengths. The results corroborate previous theories suggesting that surface effects have a major impact on melting temperature and hybridization. Their findings suggest that such surface effects reduce the melting temperature of hybrids significantly, and that probes closer to the surface need to be of higher affinity.
The data also pointed to a variation in signal between surface-proximal and surface-distal probes, with the signal from a proximally placed probe only equating that from a distally placed probe when washed at a 30-fold higher sodium ion concentration.
The upshot, according to the researchers, is that surface effects impact probe binding to a far greater extent than previously assumed. In order to achieve maximum hybridization to the complete oligo, a 60-mer polynucleotide probe, for example, should be designed as three separate sections—proximal, central, and distal. Probe segments nearer to the surface need to exhibit higher melting temperatures, whereas central and distal segments should display relatively low melting temperatures and binding strengths in order to avoid hairpins or partial hybridization.
“We are continuing to publish research demonstrating the utility of ionic gradients in the design of probes and assays,” Dr. Dufva stressed. “Additionally, our studies suggest that questions should be raised with respect to the quality of data obtained from current commercial arrays. In part, this is because the methods traditionally used to design and predict probe properties, such as nearest neighbor algorithm, may not as accurately estimate binding strength, specificity, and/or stability for immobilized probes, as previously believed.”
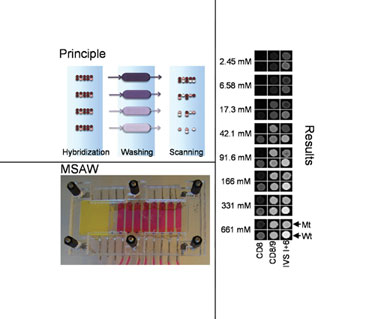
In MSAW operations, a slide with a sample is hybridized and then mounted into an MSAW, where it is washed and subsequently scanned. (Technical Univ. of Denmark)
Portability
Microarrays may be relatively inexpensive to produce, and capture several thousand items of information, but reading the fluorescence signals generated requires the use of dedicated (heavy and expensive) scanning instrumentation. This requirement has largely limited the utility of microarrays to research and drug discovery/development laboratories where such instrumentation is close at hand, said Levi Gheber, Ph.D., senior lecturer in Ben-Gurion University’s department of biotechnology engineering.
“Existing chip technologies generate spots of about 100 microns in diameter, spaced about 300–400 microns apart, and scanners are necessary because, using a standard fluorescence microscope, you can only fit in about nine spots within the field of view of a 20x objective. Yet, overcoming the need for large scanning instrumentation would make chip technology more portable and could even open the way to the development of a new generation of bioarrays for applications including point-of-care diagnostics and environmental monitoring.”
In theory, simply squeezing a greater number of much smaller spots on a slide would allow arrays to be read using a standard optical fluorescence microscope. “If an array had spots of just 1 micron in diameter rather than 100 microns, thousands would potentially be visible in one viewfield of a 20x objective lens,” Dr. Gheber continued. “The ability to generate arrays with much smaller, higher density spots would also require the use of far less biological material, and potentially allow shorter incubation times because of the reduced distances travelled by molecules within the spot.”
The situation is complicated, however, because smaller spots throw up new problems in terms of reduced signal intensity, decreased signal to noise ratio (SNR), and chip surface chemistry, before even considering the technicalities associated with accurately depositing potentially nano-sized spots of biological molecules. It’s a subject being addressed at length by the Ben-Gurion University researchers.
“Fluorescence intensity and signal to noise ratio are the most obvious hurdles,” Dr. Gheber continued. “Fluorescence signal is proportional to the area of the spot, rather than its diameter. If you try to reduce spot diameter 100-fold, from 100 microns to just 1 micron, for example, the spot area and, hence, signal intensity is actually reduced 10,000 times, while background noise stays constant. The problem is compounded further because, in fact, the reduction in signal intensity is even more dramatic due to the limited number of binding molecules per spot.
“Miniaturization must also take into account additional parameters concerning immobilization and chip-surface properties such as autofluorescence and coating homogeneity,” Dr. Gheber said. “There has, as far as we are aware, been little study of the many factors that would impact on how such tiny spots would perform.”
To investigate these parameters further, the Ben-Gurion University team devised mathematical models capable of evaluating multiple factors impacting SNR and predicting the properties of different chip surfaces. Their results suggest that, under suitable conditions, spots with diameter as low as 400 nm are detectable with an SNR above 10, at 1.3 mg/mL target molecule concentration. For spots of 1 micron diameter, detection is feasible at concentrations of just 1 ng/mL. Dr. Gheber presented results from this research at Select Biosciences’ “Advances in Microarray Technology” meeting in Stockholm.
Nano Fountain Pen Technology
The team has, in parallel, published several papers describing a nano fountain pen (NFP) technology for depositing nanoscale spots with atomic-force sensing accuracy. In addition to generating the bioarray spots, the device can also be used in the fabrication of polymer lenses that focus the fluorescence signal generated by each spot to help improve signal detection.
The ability to generate spots at the nanometer scale has already been demonstrated using dip-pen lithography (DPN), a technique that uses an atomic-force microscope probe dipped in an “ink” of biological molecules to deliver a spot on a solid support.
In contrast, the nano fountain pen technology developed by Dr. Gheber’s team uses cantilevered nanopipettes as the atomic-force microscope sensors. The heat-drawn nanopipettes have a sharp-end aperture of about 100 nm. When the capillaries are filled with the molecular ink, the liquid flows to the end of the pipette but does not flow out until the tip is contacted onto a surface.
According to Dr. Gheber, published work has already shown that the technology can be used to print a line of protein G with a width of about 500 nm and thickness of 40 nm, GFP dots of about 280 nm and thickness of 4 nm, and functional enzymes, without the requirement for specific treatment of the substrate.
The nano fountain pen technology has been exploited in the University’s development of polymer microlenses that help focus and enhance fluorescence signals generated by miniaturized spots. The microlens monomers are deposited onto the reverse of a glass microarray surface, over the area of each biological spot. UV-assisted polymerization generates spherical polymer microlenses of about four to nine microns in diameter, which focus the signal generated by each spot. The optical properties of the microlenses can also be controlled by varying the deposition time of the monomer solution.
“Our nano fountain pen and microlens platforms represent new technologies we hope will help overcome some of the technical hurdles of chip miniaturization,” Dr. Gheber noted. “Such technological capabilities must be combined with mathematical models to help accurately define optimum spot and probe parameters, together with surface chemistry. Combine these capabilities with new methods for constructing and imaging each chip in a single instrument, and the potential for using microarrays in clinical and diagnostic fields becomes evident.”
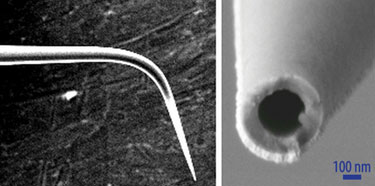
The tapered end of a nanopipette used in nano fountain pen technology. (Ben-Gurion University)
Spot Characteristics
Researchers at Cornell University’s department of applied and engineering physics have addressed the issue of microarray spot uniformity, shape, and resolution, by exploiting the principle of a childhood drawing aid—the stencil.
“Our parylene peel-array technology centers on a simple concept of a polymer stencil placed on top of the surface of the array,” explained Christine Tan, the technology’s developer. “The stencil acts as a physical constraint for spotted biomolecules, so you can deposit your molecules by whatever method you choose over the top, and then peel off the parylene layer to leave highly uniform spots. The technology can be used to generate any size, shape, or density of spot pattern.
“Some of our most recent work has even demonstrated the feasibility of generating nanometer-sized array features with nanoscale resolution comparable to existing printing technologies like dip-pen lithography. This could potentially allow the future production of nanoarrays using existing arrayjet printers or spotters, which are currently only capable of printing spots with diameters in the 10–100s of microns range.”
The flexibility of the system is inherent in its simplicity, Tan said. “The peel-arrays can be used either as stand-alone technology, to generate large uniform arrays of any biomolecule or even cells, or as a complement to any existing microarray-spotting method. The stencils act to cleanup print-spot morphology and can facilitate the printing of multiple species on a surface.
“When combined with standard DNA-printing instrumentation, for example, we have shown the reproducibility of DNA microarrays is significantly improved. By reducing the traditional DNA-drying artefacts of donut-shaped spots, we can make the standard deviations of DNA microarray experiments much smaller.”
Importantly, the peel-array technology can be used in wet or dry environments and has demonstrated major benefits in applications such as patterning lipid bilayers and proteins, where hydration is required to maintain bilayer and protein integrity, Tan added.
Additional studies with patterning fibronectin have confirmed that by using the parylene peel-arrays the protein retains its 3-D conformity and functionality for cell binding. “The parylene stencils allow users to pattern molecules and keep them hydrated. We have also used the stencils to generate uniform, large-area arrays of single tumor cells to investigate the role of cell-cell interaction in cancer, and to pattern lipid bilayers embedded with antigen for studying immune cell activation.”
Tan is looking for investors to start a business. “This is a useful technology with many potential applications in drug screening, cancer studies, biosensors, and basic science research,” she said. “We hope to make this simple and adaptable product available to everyone who uses microarrays.”
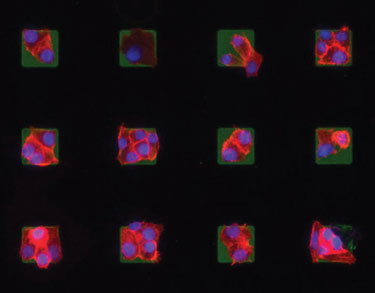
Microarrays of cells created using the parylene peel-off technology can be useful for testing drug efficacies and cancer studies.
Laser-Printing Peptide Arrays
PEPperPRINT has developed a laser-printing technology for combinatorial peptide synthesis, which it believes could bring the utility of peptide microarrays more in line with DNA microarrays in terms of array density, flexibility, and price. Spun out from the German Cancer Research Center in Heidelberg, PEPperPRINT just started commercializing its technology, offering high density peptide arrays on demand, along with iterative, high-throughput peptide-screening services for biomarker discovery and drug development.
“PEPperPRINT was founded back in 2001 to develop technologies addressing issues that were holding back the attainment of affordable, flexible high-density peptide chip manufacture,” explained CEO Volker Stadler, Ph.D., who presented the technology at the Stockholm meeting.
“At the time, we realized that existing processes that worked for the in situ synthesis of oligonucleotides such as lithographic methods weren’t as suitable for peptides. This is, at least in part, because while constructing DNA oligos requires only four monomers corresponding to the four bases, peptides are constructed from a pool of 20 amino acid monomers. Inkjet technologies are also far from ideal for printing amino acids. Liquid toner droplets of amino acids spread on the support, limiting spot density, and droplet evaporation can disturb coupling chemistry.”
PEPperPRINT’s answer was to abandon inkjets and upgrade to a laser-printing approach. The resulting instrumentation is based on a conventional color laser printer, but instead of accommodating four-color ink cartridges, the printer uses 20 amino-acid toner units.
“Our current laser-printer technology can spot at a 20-fold higher density than other state-of-the-art technologies, and by next year we hope to have developed a new system that will increase printing speed and spot density by another factor of four.”
The process itself involves mounting the surface-modified glass slide onto a linear stage that drives the slide under the printer units with micrometer accuracy. Printing the first “layer” of amino-acid toners takes just one minute. The slide is then heated, to melt all the amino-acid toners at once to intiate coupling, followed by routine washing and deprotection steps. It can then undergo the next round of printing.
PEPperPRINT believes that one of the major advantages of its laser-printer system is that the activated amino acids are embedded in solid toner particles, rather than in a liquid. This essentially immobilizes the amino acids in a stable state, so they can be stored within the toner for months without degradation, significantly reducing material costs and wastage.
“The heating process after each round of printing raises the temperature just above the melting point of the amino acids, which isn’t high enough to cause evaporation,” Dr. Stadler continued. “And because the toner matrix has an oily consistency, it doesn’t spread on the slide surface. This is a key feature that allows us to achieve much higher spot densities.”
PEPperPRINT is using the laser-printing technology to offer custom peptide chips, on both object slides and scalable glass slides. “The flexibility of the laser printing process means the cost is kept low however many chips are ordered, and whether we print a number of identical or completely different chips,” Dr. Stadler stressed. “In 2010 we aim to launch a web platform, PEPSlide®, which will allow customers to design their peptide chips on their own PC and send them through via the Internet, directly to the printing instrumentation.”
In addition to its custom chip service, PEPperPRINT also offers peptide and peptidomimetic screening services for applications including biomarker discovery, antibody/enzyme profiling, diagnostic imaging, and drug development.
“We can design individualized peptide arrays of up to 156,000 spots per slide for a wide range of screening applications. The combination of the system’s throughput, flexibility, and array density means that R&D can cost effectively use peptide arrays for sequential, iterative rounds of screening, for example to find and optimize high-affinity binders to target or probe proteins of interest.”
“Proteome research is now provided with a highly efficient tool that PEPperPRINT expects to play a similar role as DNA chips in genome research do today,” Dr. Stadler concluded.
“After starting the operative business in 2009, PEPperPRINT is going to establish both product lines, the customized peptide microarrays, and the iterative high-throughput screening routine, in the market in 2010. The next steps will include pilot studies with selected pharma companies and the launch of PEPSlide for the global web-based distribution of peptide chips.”

The inside of PEPperPRINT’s peptide laser printer with drives for the printing gears







