January 15, 2014 (Vol. 34, No. 2)
As epigenetics is unveiling new layers of organization and regulation, it is becoming increasingly clear how little we knew about fundamental processes shaping biological systems.
Many transformative changes from virtually every biomedical discipline owe their existence to epigenetics. The development of new technologies and the refinement of the existing ones are marking an era when, in addition to introducing new concepts, we are also revisiting, and often reshaping, many of the existing ones.
Over 1,300 genome-wide association studies have been published since 2005. “One of the big challenges with GWAS is that there is always the concept of missing heritability,” says Peter C. Scacheri, Ph.D., associate professor of genetics and genome sciences at Case Western Reserve University.
Missing heritability refers to the fact that, for most common diseases, even after all known genetic risk factors are taken into consideration, collectively they do not explain all the heritability. For example, even though at least 42 different genes were implicated in the risk to develop type 2 diabetes mellitus, collectively they only explain approximately 20% of the heritability.
“Understanding missing heritability has been a major challenge in the field,” notes Dr. Scacheri. Some of the current views are that additional genetic variants, not yet discovered, may exist, or that epigenetic changes may hold the explanation. “It is also quite possible that the way that we have been going about GWASs to identify regions that confer genetic risk has not been done as well as it could be,” adds Dr. Scacheri.
Of the several hundred SNPs that were uncovered and linked to complex diseases, over 90% are located in noncoding regions of the genome, and many map within enhancer elements. By focusing on six autoimmune conditions, Dr. Scacheri and colleagues revealed that several SNPs in a given cell type map to multiple enhancer elements, cooperatively influencing gene expression in a model that they named the “multiple enhancer variant hypothesis.”
“We showed that many genetic loci linked to various common diseases not only localize within enhancer elements in a given cell type, but also that these combinations of genetic variants are probably contributing to the effects of gene expression and confer the risk for human disease,” asserts Dr. Scacheri.
This finding provides a new model to explain the ability of noncoding genetic variants to shape gene expression. “The way investigators should be looking for loci that confer risk to a given disease is by starting with the epigenetic marks and the chromatin maps, and using these as a basis for GWASs,” says Dr. Scacheri. Many common diseases have genetic and epigenetic components, and it is still unclear how much of the epigenetics can explain common diseases, but an intricate and multilayered interplay between genetic and epigenetic elements has increasingly been unveiled with many approaches.
Technological advances allowed investigators to study the biology of epigenetic modifications in ways that have not been available in the recent past. “But one of the challenges is that much of what has been done is still in cell lines. As the next step, we should map out epigenetic profiles in primary cells and tissues derived from the human body,” observes Dr. Scacheri.
This endeavor promises even more challenges, as every tissue has a number of distinct cell types, and epigenetic profiles are known to vary not only across cell types, but also among cells of the same type. “We think that trying to sort out the epigenetic landscape in all those specific cell types is possible, but right now it seems a little bit daunting,” concludes Dr. Scacheri.
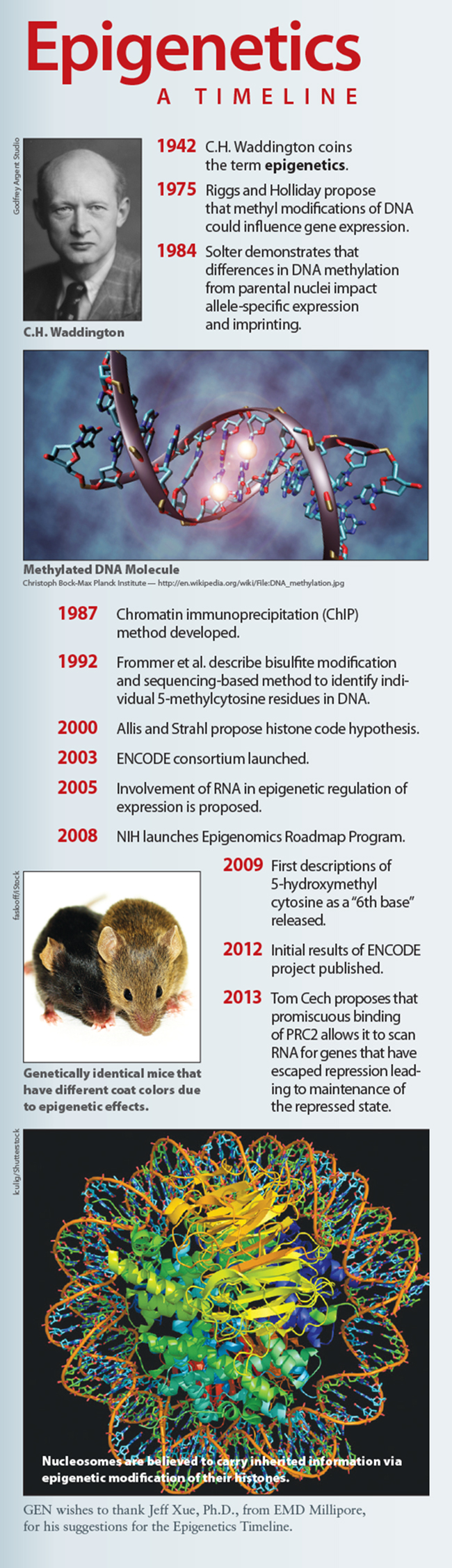
Epigenetics: A Timeline
Markers of Disease Progression
“The current challenge in dermatology is differentiating benign melanocytic nevi from primary or dysplastic melanoma, which represent the early stages of malignant differentiation,” says Ranjan J. Perera, Ph.D., associate professor and scientific director, analytical genomics and bioinformatics, at Sanford-Burnham Medical Research Institute at Lake Nona. Significant efforts in Dr. Perera’s lab are focusing on the development of markers for the early detection of malignant melanoma.
The incidence of melanoma has been steadily rising in the United States for several decades, and if diagnosed early, melanoma is curable in virtually all patients, but early diagnosis is challenging. Several million biopsies are performed annually in the United States for suspicious melanoma, and they are examined by immunohistochemistry, but variables related to the technique, the microscope, and the staining procedure affect the results.
“We are trying to add more specificity and sensitivity to the existing biomarkers,” notes Dr. Perera. He and his colleagues are focusing on developing genome-level epigenetic molecular markers that can diagnose melanoma at the earliest stages. By using next-generation sequencing, Dr. Perera and colleagues examined the genome-wide distribution of methylated CpG islands from coding and noncoding RNA genes in melanoma-derived cell lines, melanoma samples, melanocytic nevi, and normal melanocytes.
This strategy unveiled specific CpG island methylation signatures that are characteristic for melanocytes and for early and late melanoma. A progressive hypomethylation of large CpG island stretches was visualized in the early stages, which corresponds to the progression to early melanoma, while a subsequent stage, the development of late-stage melanoma, was accompanied by extensive hypermethylation. By integrating the profiles obtained after pharmacological demethylation with data from RNA-Seq experiments, Dr. Perera and colleagues described specific signatures encompassing a co-expression network of differentially expressed genes at specific stages during cancer progression.
This analysis, which mapped over 19,000 CpG sequences, illustrates the dynamic CpG methylation pattern that marks melanoma development and progression, and points toward the potential to reclassify melanomas based on their gene expression and epigenetic signatures, and to develop biomarkers. “We would actually be able to detect and obtain this information by noninvasive diagnosis, such as in serum, blood, or urine.”
Previously, Dr. Perera and colleagues were the first ones to unveil the very active role that microRNA-211 plays in regulating large numbers of genes in melanoma. Work completed since then by several groups that used next-generation sequencing revealed that microRNA-211 is the most differentially regulated microRNA in melanoma and can be used as a diagnostic marker. “The metastasis of melanoma is very fast, and the current challenge, which is finding a marker to noninvasively discriminate malignant from benign lesions in very early stages, could be lifesaving,” concludes Dr. Perera.
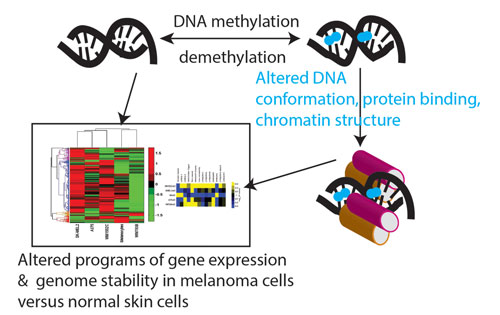
Researchers at the Sanford-Burnham Medical Research Institute are developing genome-level epigenetic molecular markers for the early detection of malignant melanoma. Their technique involves integrating the CpG patterns obtained after pharmacologic demethylation with data from RNA-Seq experiments.
Studying the Methylome
“We study a large group of enzymes, the family of lysine methyltransferases,” says Or Gozani, M.D., Ph.D., associate professor of biology at Stanford University. The human genome encodes over 50 candidate lysine methyltransferases, enzymes that are able to add one, two, or three methyl groups to lysine residues on histones and other proteins.
The biological function of many members of this family is not yet known, but several representatives were implicated in malignancies, immune disorders, and critical processes such as stem cell maintenance. “We wanted to develop an approach to de-orphanize these enzymes and study their biology,” notes Dr. Gozani.
While antibodies have provided the opportunity to survey several types of histone post-translational modifications, this has been incomparably more challenging in the case of lysine methylation. “We had to develop an alternative approach, one that relies on reagents that do not require antibodies,” says Dr. Gozani.
This strategy involved generating an engineered affinity reagent that is highly specific for mono- and dimethylated lysine residues and can recognize and bind them, irrespective of the surrounding sequence. This reagent was based on the three repeats of the malignant brain tumor (MBT) domain of L3MBTL1, which plays a key role in chromosomal compaction.
Each 100-amino-acid MBT repeat adopts a structure that resembles a three-leaved propeller, with a central cavity in the second MBT repeat that binds mono- and dimethylated lysine residues. The fact that the 3xMBT domain does not establish significant contacts with the side chains of surrounding amino groups ensures the sequence-independent context of the recognition.
“This was the first time detection of lysine methylation was performed at the proteomic level,” asserts Dr. Gozani. By using this reagent in a proteome pull-down experiment, graduate student Katie Moore and post-doctoral fellow Dr. Scott Carlson in Dr. Gozani’s group, together with colleagues, were able to enrich for over 300 proteins and, for a subset of those, to map methylated lysine residues, leading to the identification of potentially several hundred methylated histones.
“We think that we captured only a fraction of the full methylome,” says Dr. Gozani. Superior reagents and better instruments will make it possible to more thoroughly investigate the full ensemble of methylated proteins. “One of the biggest challenges is being able to study proteins that are not abundantly methylated, and understanding the consequences of the individual changes at the systems level will be another major challenge,” comments Dr. Gozani.
Imprinted Genes
A special subset of genes known as imprinted genes is attracting increasing attention. Imprinting is an epigenetic process that involves the monoallelic, parent-of-origin-specific silencing of specific genes. Both DNA methylation and histone post-translational modification contribute to the establishment of the imprintome, the functional definition coined in 2009 that refers to the totality of the genomic regions that control the monoallelic silencing of imprinted genes.
“Without understanding imprinting, I do not think we will ever truly understand neurological disorders, such as schizophrenia and autism,” says Randy L. Jirtle, Ph.D., professor of epigenetics at the University of Bedfordshire and visiting professor at the University of Wisconsin-Madison. In a recent study that examined DNA from umbilical cord blood leukocytes, Dr. Jirtle and colleagues revealed that paternal obesity is inversely related to the increased methylation of differentially methylated regions in the imprinted insulin-like growth factor 2 (IGF2) gene, suggesting that paternal obesity impacts offspring health in a transgenerational manner.
Dr. Jirtle and colleagues were the first ones to show that the process of imprinting evolved approximately 150 million years ago in a common ancestor to Therian mammals. One of the main reasons to focus on imprinted genes is that, due to their monoallelic expression, they are functionally haploid. As a result, the impact of any perturbations that change the expression pattern of imprinted genes would be dominant. In human somatic cells, some imprinted genes are of maternal origin while others are of paternal origin. Importantly, a number of human diseases are linked to the epigenetically altered expression of imprinted genes.
A thought-provoking aspect related to imprinted genes is that imprinting does not translate well across species, opening significant difficulties in terms of extrapolating findings from animal models. Mouse models were instrumental in revealing general principles, such as the ability of environmental factors to epigenetically shape gene expression. Several years ago, using the agouti viable yellow mouse model, Dr. Jirtle and colleagues showed that maternal nutrition modulates DNA methylation and shapes the phenotype of the offspring, opening the era of environmental epigenomics.
“Nutritional influences will probably alter the epigenome and the phenotype in humans, too, but one should not extrapolate from mice to humans, because the imprintome varies greatly across species, and even if imprinted genes were the same between two species, they can still be regulated differently,” cautions Dr. Jirtle. An illustration of challenges to come, and potential pitfalls, is provided by the example of the IGF2 gene, which is imprinted and paternally expressed in both mice and humans, but is markedly differently regulated after birth in the two species.
Additional considerations suggest that imprinted genes pose complexities unlike the ones encountered in most other chapters of biology. For example, significant increases in the intergenic distances are seen in human, but not mouse imprinted genes. Also, imprinted genes present in mice are not seen in humans.
“There are places for mice in model systems, but it can be difficult to extrapolate epigenetic effects even to different mouse strains, let alone humans,” observes Dr. Jirtle.
In mice, the impact of deleting functional copies of imprinted genes depends on whether the maternally or the paternally inherited allele is the one that was affected. Deleting the paternally inherited Igf2 gene results in viable, but smaller progeny, whereas deleting the maternal copy of the H19/Igf2 imprint regulatory region results in increased Igf2 expression and larger progeny. In humans, deleting a chromosomal region on the long arm of chromosome 13 leads to the Prader-Willi syndrome when the paternally expressed allele is deleted, or to the Angelman syndrome when the maternally expressed allele is deleted.
Certain genes exhibit monoallelic expression but, unlike in the preceding examples, silencing does not occur in a parent-of-origin-dependent manner. Instead, the maternally inherited allele is expressed in one individual, and the paternally inherited allele in another one. “We identified two such genes in the genomic region that results in DiGeorge syndrome when deleted,” asserts Dr. Jirtle.
This inheritance is less well understood than genomic imprinting, because gene expression, even though still monoallelic, is not parent-of-origin-dependent. Rather, it appears to be individual-dependent, and understanding the mechanisms by which this occurs promises to be considerably more time-consuming and challenging. “Genomically imprinted genes are only a subset of those that are monoallelically expressed, but defining them and how they are regulated is fundamentally essential to understand human disease,” concludes Dr. Jirtle.
The Ticking of the Epigenetic Clock
The legend says that when Eos, the goddess of the dawn, asked Zeus to grant immortality to her lover Tithonus, she failed to also ask for his eternal youth. According to some stories, after he grew old and became weak, she transformed him into a grasshopper, which in Greek mythology is immortal. As tales from many cultures illustrate, understanding why and how we age, along with preventing, slowing, or reversing this process, are topics that accompanied and fascinated societies since the earliest times.
“Aging is not well understood and, in addition, we do not have accurate ways to measure and quantitate the aging process,” says Steve Horvath, Ph.D., professor of genetics and biostatistics at the University of California, Los Angeles. Multiple lines of experimental evidence, along with observational studies, increasingly point toward the importance of epigenetic factors in shaping the process of aging.
In an effort to develop a quantitative assessment tool to express the age of tissues from an epigenetic perspective, Dr. Horvath used publicly available DNA methylation datasets encompassing samples from 51 different organs, tissues, and cell types, and examined the methylation of 353 CpG dinucleotides. “This epigenetic clock is the most accurate biological clock that I am aware of,” observes Dr. Horvath.
A salient characteristic of this approach, which can be applied to study epigenetic changes that accompany development, aging, and disease, is that it can be used for virtually any cell type. “The research community should carefully evaluate whether this epigenetic clock could be used to detect or evaluate developmental disorders,” cautions Dr. Horvath. The possibility differentially quantitating accelerated aging across tissues, as part of specific diseases, promises to add a new layer of interrogation to understanding disease pathogenesis.
By examining this dataset, Dr. Horvath revealed that the epigenetic clock is perturbed in several circumstances. “Cancer very much disrupts the epigenetic clock,” emphasizes Dr. Horvath. The analysis revealed that certain tissues looked systematically older, while others looked younger than expected based on the chronological age. Estrogen- or progesterone-receptor-positive breast cancers appeared to be, in this analysis, much older than other breast cancer types.
“We need to understand why does one tissue look older and the other one younger, because this will, among other things, help up understand why we age,” assert Dr. Horvath. The study of epigenetic age could extend beyond humans, because in several chimpanzee tissues, the DNA methylation age was similarly correlated with the chronological age. Based on the current data, this approach appears to be of more limited value in gorillas, a potential reflection of their more remote evolutionary age.
A significant limitation, at this time, is the lack of longitudinal data to portray the changes that occur over time in the epigenetic clock. This perspective promises to be particularly informative, considering that data from cord blood unveiled detectable changes as soon as one year after birth. “The most acute need, at this time, is to develop such an epigenetic clock in a mouse model, to help us understand what biological processes are measured by this clock,” concludes Dr. Horvath.
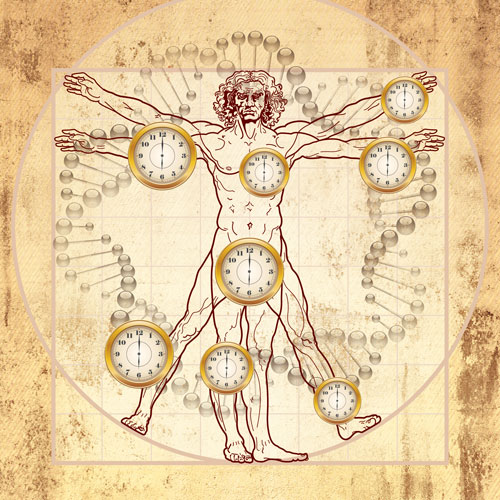
By quantitatively assessing the age of tissues from an epigenetic perspective, researchers at the University of California, Los Angeles, have essentially created an epigenetic clock. It raises the possibility of using epigenetics to measure aging or disease in one tissue versus aging or disease in another.



